|
|
|
|
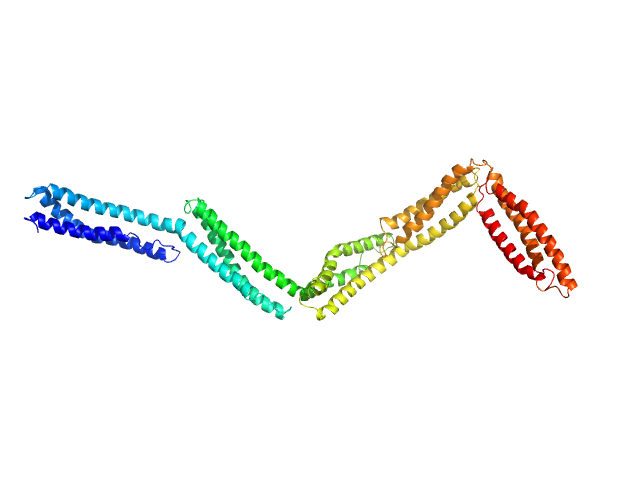
|
| Sample: |
Dystrophin (R11-15 human dystrophin fragment) monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% v/v D2O, pH: 7.1
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 1
|
How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids.
J Struct Biol :107411 (2019)
Mias-Lucquin D, Dos Santos Morais R, Chéron A, Lagarrigue M, Winder SJ, Chenuel T, Pérez J, Appavou MS, Martel A, Alviset G, Le Rumeur E, Combet S, Hubert JF, Delalande O
|
| RgGuinier |
6.2 |
nm |
| Dmax |
27.4 |
nm |
| VolumePorod |
146 |
nm3 |
|
|
|
|
|
|
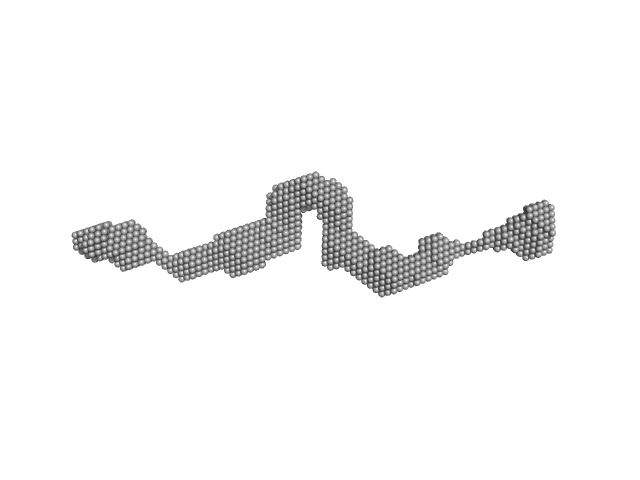
|
| Sample: |
Dystrophin (R11-15 human dystrophin fragment) monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% v/v D2O, pH: 7.1
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 1
|
How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids.
J Struct Biol :107411 (2019)
Mias-Lucquin D, Dos Santos Morais R, Chéron A, Lagarrigue M, Winder SJ, Chenuel T, Pérez J, Appavou MS, Martel A, Alviset G, Le Rumeur E, Combet S, Hubert JF, Delalande O
|
| RgGuinier |
6.2 |
nm |
| Dmax |
28.1 |
nm |
| VolumePorod |
144 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dystrophin (R11-15 human dystrophin fragment) monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% v/v D2O, pH: 7.1
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 1
|
How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids.
J Struct Biol :107411 (2019)
Mias-Lucquin D, Dos Santos Morais R, Chéron A, Lagarrigue M, Winder SJ, Chenuel T, Pérez J, Appavou MS, Martel A, Alviset G, Le Rumeur E, Combet S, Hubert JF, Delalande O
|
| RgGuinier |
8.1 |
nm |
| Dmax |
29.6 |
nm |
| VolumePorod |
231 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human dystrophin central domain R8-15 fragment monomer, 100 kDa protein
|
| Buffer: |
NaP 10 mM, NaCl 500 mM, EDTA 1 mM, Glycerol 2%, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Sep 23
|
How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids.
J Struct Biol :107411 (2019)
Mias-Lucquin D, Dos Santos Morais R, Chéron A, Lagarrigue M, Winder SJ, Chenuel T, Pérez J, Appavou MS, Martel A, Alviset G, Le Rumeur E, Combet S, Hubert JF, Delalande O
|
| RgGuinier |
10.1 |
nm |
| Dmax |
36.0 |
nm |
|
|
|
|
|
|
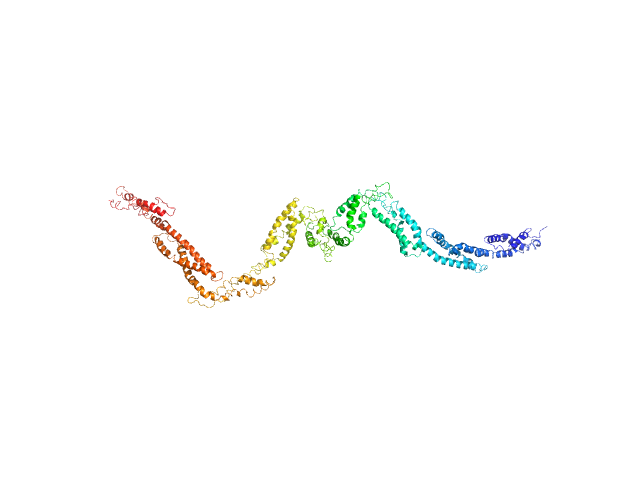
|
| Sample: |
Human dystrophin central domain R11-19 fragment monomer, 117 kDa protein
|
| Buffer: |
NaP 20 mM, NaCl 300 mM, EDTA 1 mM, Glycerol 2%, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 May 11
|
How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids.
J Struct Biol :107411 (2019)
Mias-Lucquin D, Dos Santos Morais R, Chéron A, Lagarrigue M, Winder SJ, Chenuel T, Pérez J, Appavou MS, Martel A, Alviset G, Le Rumeur E, Combet S, Hubert JF, Delalande O
|
| RgGuinier |
8.8 |
nm |
| Dmax |
37.5 |
nm |
| VolumePorod |
513 |
nm3 |
|
|
|
|
|
|
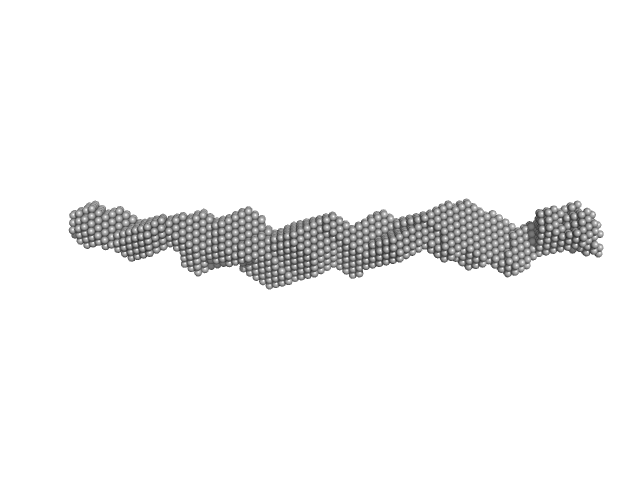
|
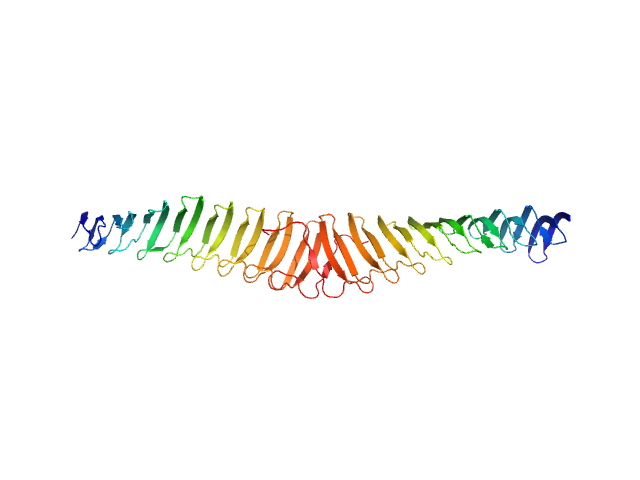
| Sample: |
Trypanosoma brucei Membrane Occupation and Recognition Nexus MORN repeats 2-15 dimer, 78 kDa Trypanosoma brucei protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jan 27
|
Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
PLoS One 15(12):e0242677 (2020)
Sajko S, Grishkovskaya I, Kostan J, Graewert M, Setiawan K, Trübestein L, Niedermüller K, Gehin C, Sponga A, Puchinger M, Gavin AC, Leonard TA, Svergun DI, Smith TK, Morriswood B, Djinovic-Carugo K
|
| RgGuinier |
6.5 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
187 |
nm3 |
|
|
|
|
|
|
|
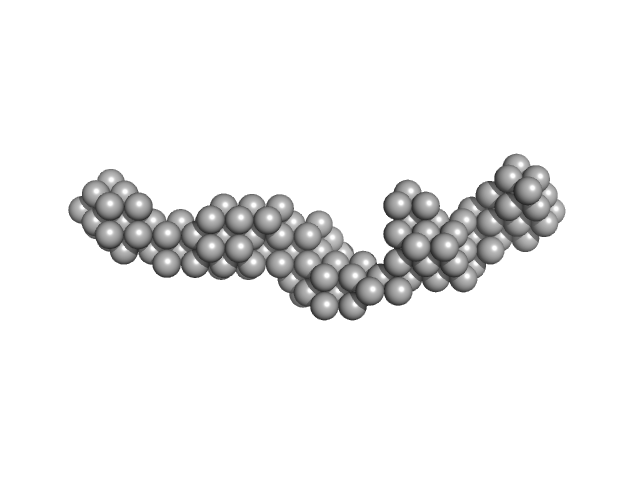
| Sample: |
Trypanosoma brucei Membrane Occupation and Recognition Nexus MORN (7-15) dimer, 52 kDa Trypanosoma brucei protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 2% (w/v) glycerol, 0.5 mM DTT, pH: 8.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 11
|
Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
PLoS One 15(12):e0242677 (2020)
Sajko S, Grishkovskaya I, Kostan J, Graewert M, Setiawan K, Trübestein L, Niedermüller K, Gehin C, Sponga A, Puchinger M, Gavin AC, Leonard TA, Svergun DI, Smith TK, Morriswood B, Djinovic-Carugo K
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
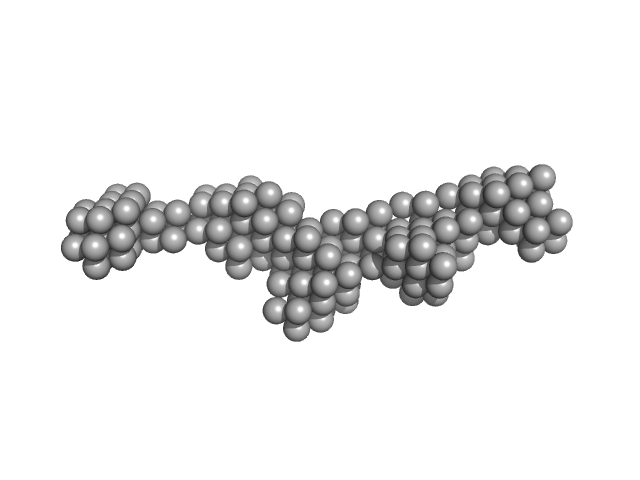
| Sample: |
Toxoplasma gondii Membrane Occupation and Recognition Nexus (MORN1) repeats 7-15 dimer, 49 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 10
|
Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
PLoS One 15(12):e0242677 (2020)
Sajko S, Grishkovskaya I, Kostan J, Graewert M, Setiawan K, Trübestein L, Niedermüller K, Gehin C, Sponga A, Puchinger M, Gavin AC, Leonard TA, Svergun DI, Smith TK, Morriswood B, Djinovic-Carugo K
|
| RgGuinier |
4.0 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
|
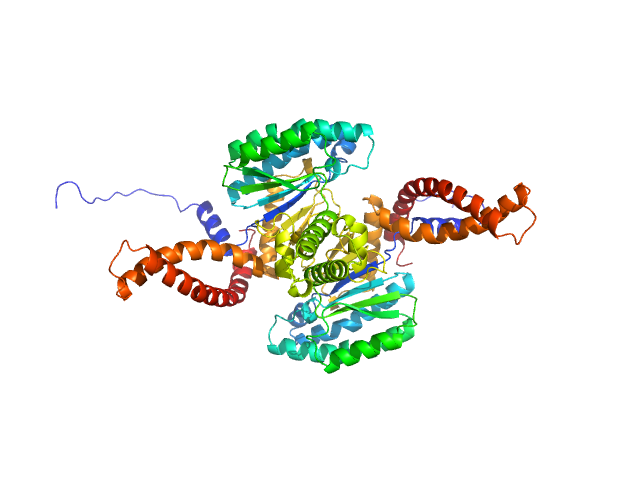
| Sample: |
MORN repeat-containing protein 1 dimer, 46 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 2
|
Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
PLoS One 15(12):e0242677 (2020)
Sajko S, Grishkovskaya I, Kostan J, Graewert M, Setiawan K, Trübestein L, Niedermüller K, Gehin C, Sponga A, Puchinger M, Gavin AC, Leonard TA, Svergun DI, Smith TK, Morriswood B, Djinovic-Carugo K
|
| RgGuinier |
3.8 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dehydrodolichyl diphosphate synthase complex subunit DHDDS dimer, 78 kDa Homo sapiens protein
|
| Buffer: |
Tris-HCl, 150 mM NaCl, 20 mM 2-mercaptoethanol and 0.02% Triton-X100, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 2
|
Structural Characterization of Full-Length Human Dehydrodolichyl Diphosphate Synthase Using an Integrative Computational and Experimental Approach.
Biomolecules 9(11) (2019)
Lisnyansky Bar-El M, Lee SY, Ki AY, Kapelushnik N, Loewenstein A, Chung KY, Schneidman-Duhovny D, Giladi M, Newman H, Haitin Y
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
122 |
nm3 |
|
|