|
|
|
|
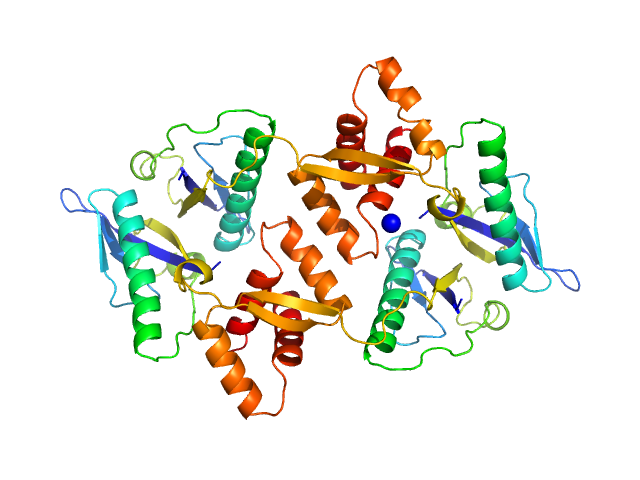
|
| Sample: |
Type II toxin-antitoxin system HicB family antitoxin tetramer, 63 kDa Burkholderia pseudomallei protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 28
|
The molecular basis of protein toxin HicA-dependent binding of the protein antitoxin HicB to DNA.
J Biol Chem (2018)
Winter AJ, Williams C, Isupov MN, Crocker H, Gromova M, Marsh P, Wilkinson OJ, Dillingham MS, Harmer NJ, Titball RW, Crump MP
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
112 |
nm3 |
|
|
|
|
|
|
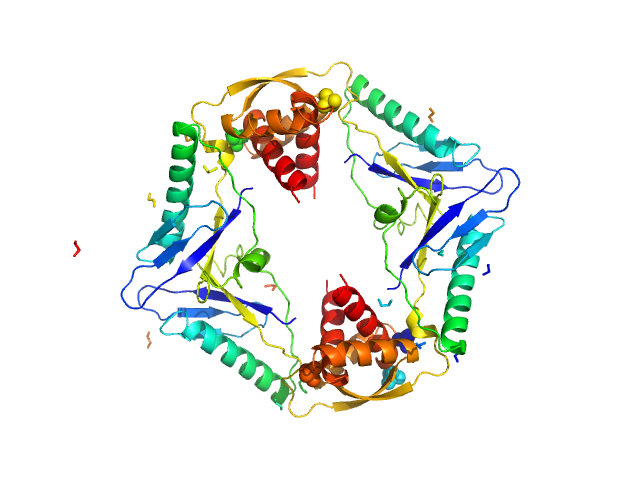
|
| Sample: |
Type II toxin-antitoxin system HicB family antitoxin tetramer, 63 kDa Burkholderia pseudomallei protein
Addiction module toxin, HicA monomer, 7 kDa Burkholderia pseudomallei protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 27
|
The molecular basis of protein toxin HicA-dependent binding of the protein antitoxin HicB to DNA.
J Biol Chem (2018)
Winter AJ, Williams C, Isupov MN, Crocker H, Gromova M, Marsh P, Wilkinson OJ, Dillingham MS, Harmer NJ, Titball RW, Crump MP
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
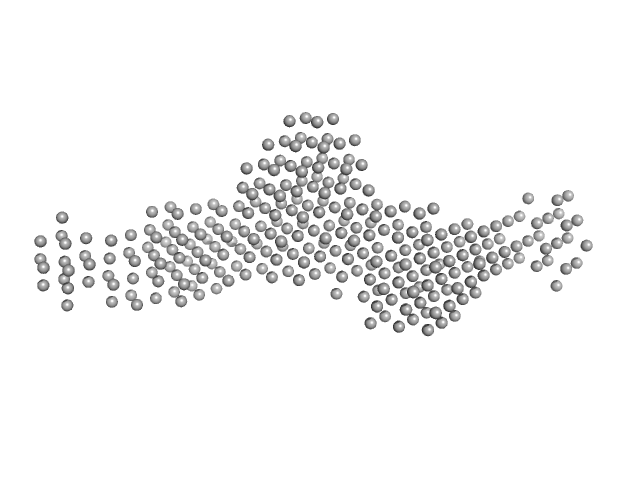
|
| Sample: |
SGT protein dimer, 92 kDa Leishmania braziliensis protein
|
| Buffer: |
20 mM Potassium Phosphate, 100 mM KCl, 10 mM EDTA, 1 mM B-mercaptoethanol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2014 May 14
|
Structural and functional studies of the Leishmania braziliensis SGT co-chaperone indicate that it shares structural features with HIP and can interact with both Hsp90 and Hsp70 with similar affinitie...
Int J Biol Macromol 118(Pt A):693-706 (2018)
Coto ALS, Seraphim TV, Batista FAH, Dores-Silva PR, Barranco ABF, Teixeira FR, Gava LM, Borges JC
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.0 |
nm |
|
|
|
|
|
|
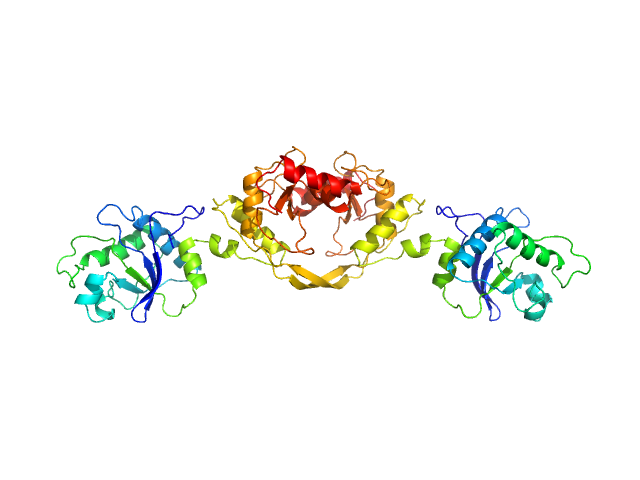
|
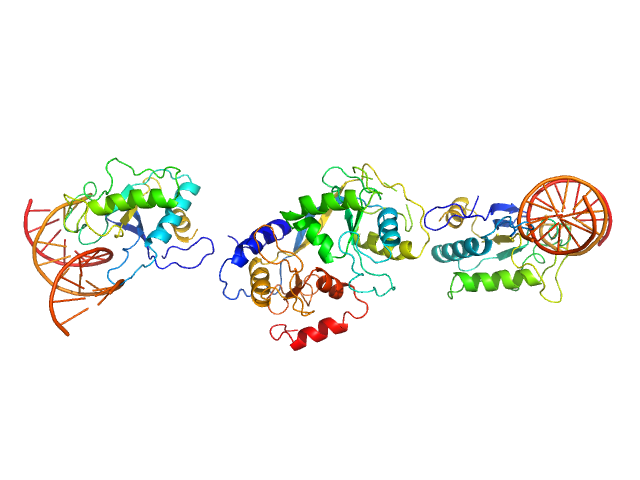
| Sample: |
5-methylcytosine-specific restriction enzyme A dimer, 65 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|
|
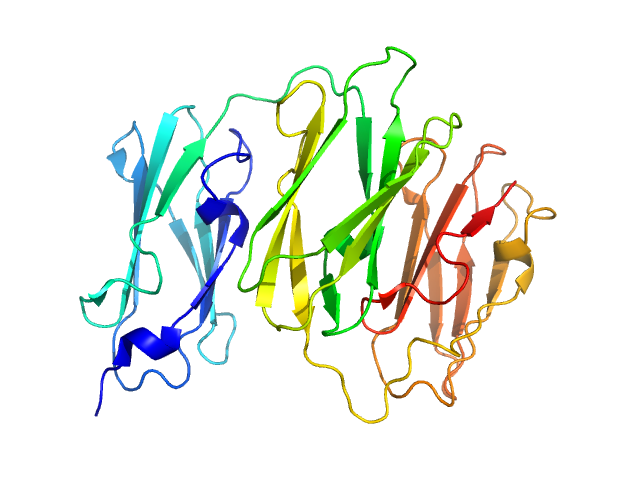
| Sample: |
cognate hemimethylated 12-bp oligoduplex dimer, 15 kDa DNA
5-methylcytosine-specific restriction enzyme A dimer, 65 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
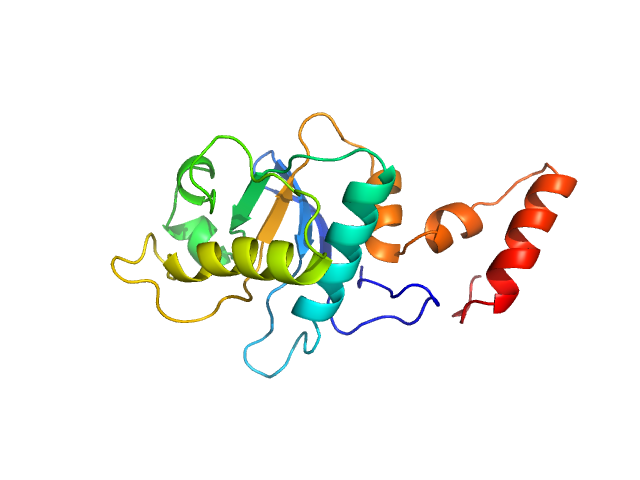
| Sample: |
5-methylcytosine-specific restriction enzyme A (N-terminal domain) monomer, 21 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
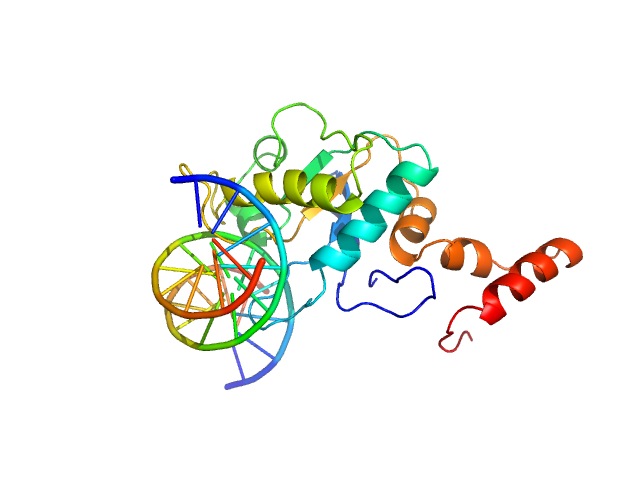
|
| Sample: |
5-methylcytosine-specific restriction enzyme A (N-terminal domain) monomer, 21 kDa Escherichia coli protein
cognate hemimethylated 12-bp oligoduplex monomer, 7 kDa DNA
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Filamin A Ig.like domains 3-5 P637Q mutant monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 10
|
Non-syndromic Mitral Valve Dysplasia Mutation Changes the Force Resilience and Interaction of Human Filamin A.
Structure (2018)
Haataja TJK, Bernardi RC, Lecointe S, Capoulade R, Merot J, Pentikäinen U
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
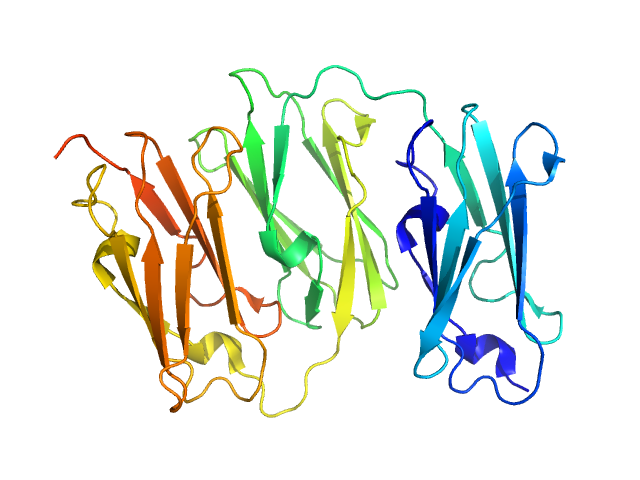
|
| Sample: |
Filamin A Ig-like domains 3-5 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 10
|
Non-syndromic Mitral Valve Dysplasia Mutation Changes the Force Resilience and Interaction of Human Filamin A.
Structure (2018)
Haataja TJK, Bernardi RC, Lecointe S, Capoulade R, Merot J, Pentikäinen U
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
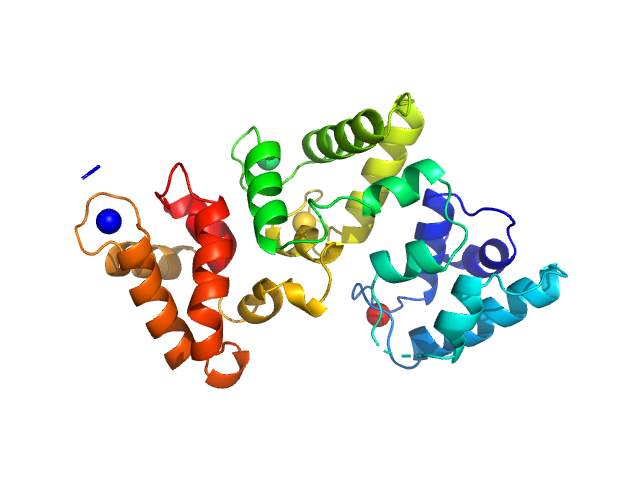
|
| Sample: |
Calbindin monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3 mM CaCl2, pH: 7.8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Feb 28
|
The X-ray structure of human calbindin-D28K: an improved model.
Acta Crystallogr D Struct Biol 74(Pt 10):1008-1014 (2018)
Noble JW, Almalki R, Roe SM, Wagner A, Duman R, Atack JR
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
47 |
nm3 |
|
|