|
|
|
|
|
| Sample: |
Integrin beta-4 (1436-1666) R1475A monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 26
|
Integrin α6β4 Recognition of a Linear Motif of Bullous Pemphigoid Antigen BP230 Controls Its Recruitment to Hemidesmosomes.
Structure 27(6):952-964.e6 (2019)
Manso JA, Gómez-Hernández M, Carabias A, Alonso-García N, García-Rubio I, Kreft M, Sonnenberg A, de Pereda JM
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Integrin beta-4 (1436-1666) R1542E monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 26
|
Integrin α6β4 Recognition of a Linear Motif of Bullous Pemphigoid Antigen BP230 Controls Its Recruitment to Hemidesmosomes.
Structure 27(6):952-964.e6 (2019)
Manso JA, Gómez-Hernández M, Carabias A, Alonso-García N, García-Rubio I, Kreft M, Sonnenberg A, de Pereda JM
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Integrin beta-4 (1436-1666) R1542A monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 26
|
Integrin α6β4 Recognition of a Linear Motif of Bullous Pemphigoid Antigen BP230 Controls Its Recruitment to Hemidesmosomes.
Structure 27(6):952-964.e6 (2019)
Manso JA, Gómez-Hernández M, Carabias A, Alonso-García N, García-Rubio I, Kreft M, Sonnenberg A, de Pereda JM
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Integrin beta-4 (1436-1666) T1663R monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 26
|
Integrin α6β4 Recognition of a Linear Motif of Bullous Pemphigoid Antigen BP230 Controls Its Recruitment to Hemidesmosomes.
Structure 27(6):952-964.e6 (2019)
Manso JA, Gómez-Hernández M, Carabias A, Alonso-García N, García-Rubio I, Kreft M, Sonnenberg A, de Pereda JM
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
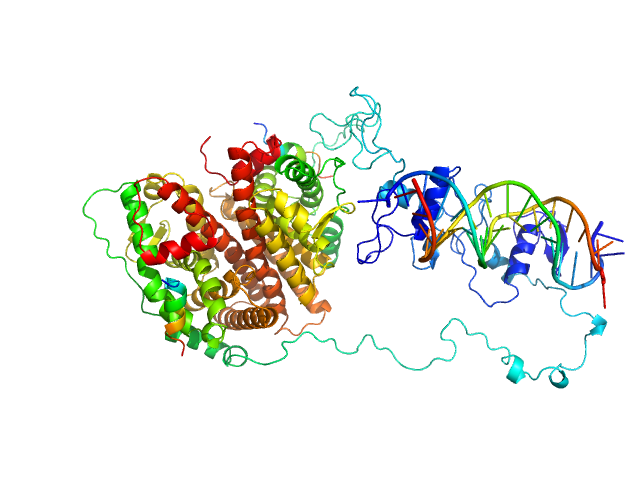
|
| Sample: |
Estrogen receptor dimer, 85 kDa protein
ERE1 monomer, 6 kDa Homo sapiens DNA
ERE2 monomer, 6 kDa Homo sapiens DNA
Estradiol dimer, 0 kDa
hERa peptide1 monomer, 2 kDa protein
hERa peptide2 monomer, 2 kDa protein
|
| Buffer: |
10 mM CHES (pH9.5), 125 mM NaCl, 5mM KCl, 4 mM MgCl2, 50 mM arginine, 50 mM glutamate, 5 mM TCEP, 5% glycerol, 10 µm Zn acetate, 10 µM estradiol, pH: 9.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Aug 10
|
Multidomain architecture of estrogen receptor reveals interfacial cross-talk between its DNA-binding and ligand-binding domains.
Nat Commun 9(1):3520 (2018)
Huang W, Peng Y, Kiselar J, Zhao X, Albaqami A, Mendez D, Chen Y, Chakravarthy S, Gupta S, Ralston C, Kao HY, Chance MR, Yang S
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
|
|
|
|
|
|
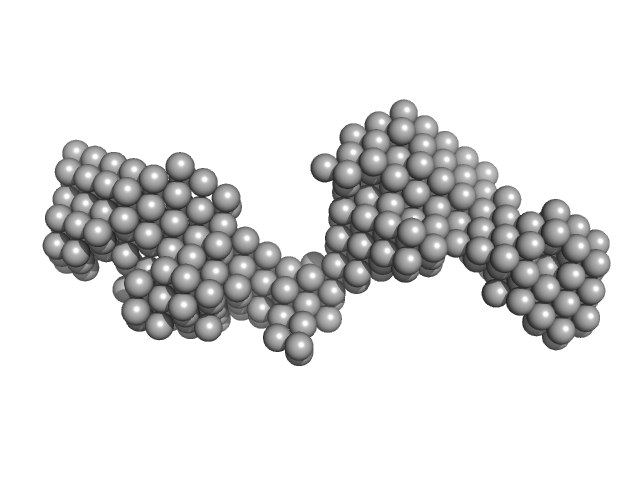
|
| Sample: |
Tegument protein UL37 monomer, 49 kDa Suid alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Inner tegument protein monomer, 62 kDa Human alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
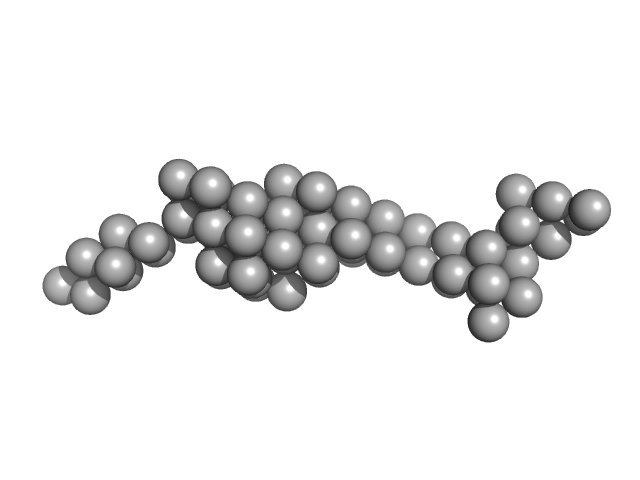
|
| Sample: |
Tegument protein UL37 monomer, 43 kDa Suid alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
|
|
|
|
|
|
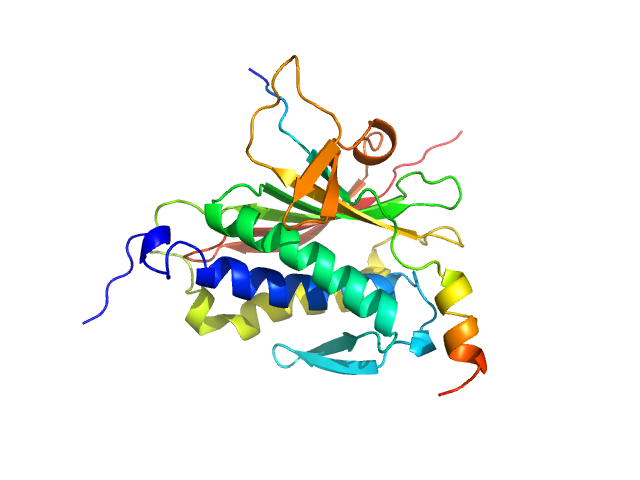
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B monomer, 24 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol, pH: 8.4
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B monomer, 24 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol, pH: 8.4
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
47 |
nm3 |
|
|