|
|
|
|
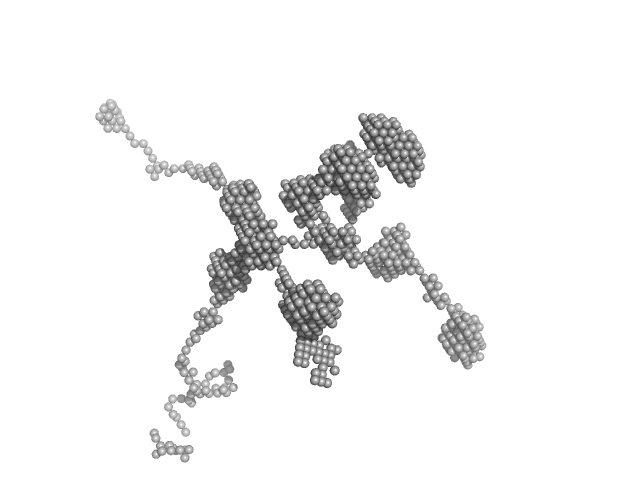
|
| Sample: |
self-amplifying RNA monomer, 3030 kDa RNA
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 10
|
Compact polyethylenimine-complexed mRNA vaccines
Nature Nanotechnology (2025)
Moreno Herrero J, Stahl T, Erbar S, Maxeiner K, Schlegel A, Bacic T, Schumacher J, Cavalcanti L, Schroer M, Svergun D, Sahin U, Haas H
|
| RgGuinier |
90.5 |
nm |
| Dmax |
200.0 |
nm |
|