|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (E121C, C162T, C552S, N556C) octamer, 566 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8
|
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jul 15
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
...Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
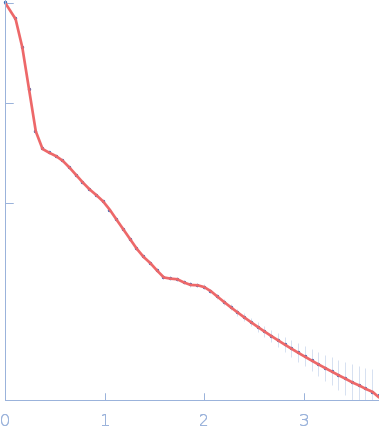
| RgGuinier |
10.3 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
1170 |
nm3 |
|