UniProt ID: P9WMU1 (2-60) Cell wall synthesis protein Wag31
|
|
|
|
| Sample: |
Cell wall synthesis protein Wag31 tetramer, 34 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
20 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 3
|
Structural basis of self-assembly in the lipid-binding domain of mycobacterial polar growth factor Wag31
IUCrJ 7(4):767-776 (2020)
Choukate K, Chaudhuri B
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
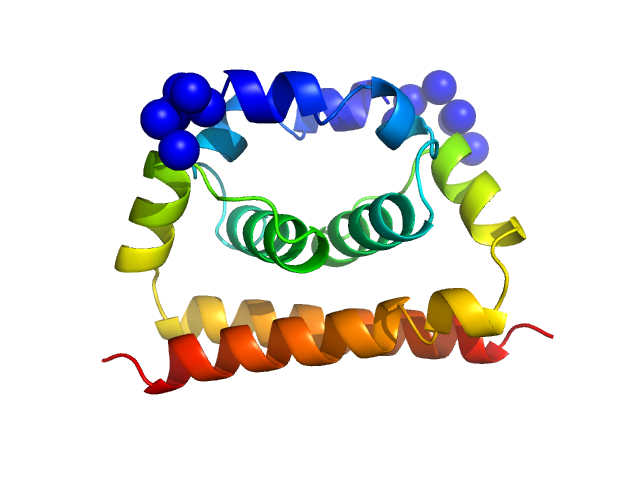
UniProt ID: P14340 (21-100) Dengue Virus 2 New Guinea C
|
|
|
|
| Sample: |
Dengue Virus 2 New Guinea C dimer, 19 kDa Dengue virus 2 protein
|
| Buffer: |
100 mM NaCl, 25 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2018 Jul 5
|
A cocrystal structure of dengue capsid protein in complex of inhibitor.
Proc Natl Acad Sci U S A 117(30):17992-18001 (2020)
Xia H, Xie X, Zou J, Noble CG, Russell WK, Holthauzen LMF, Choi KH, White MA, Shi PY
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
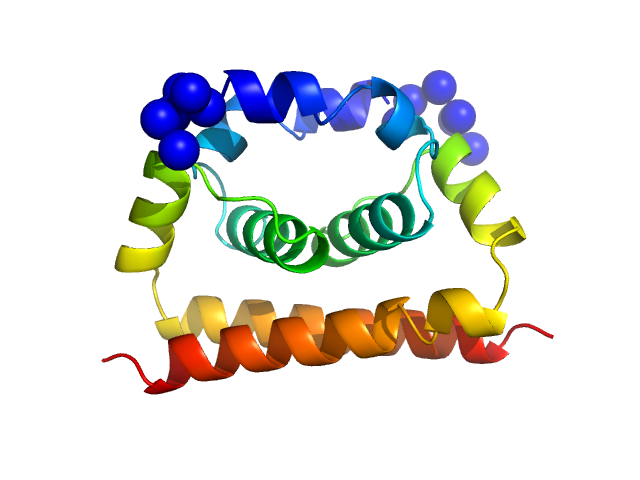
UniProt ID: P14340 (21-100) Dengue Virus 2 New Guinea C
|
|
|
|
| Sample: |
Dengue Virus 2 New Guinea C dimer, 19 kDa Dengue virus 2 protein
|
| Buffer: |
100 mM NaCl, 25 mM HEPES, pH 7.4, 10 µM ST148, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2018 Jul 26
|
A cocrystal structure of dengue capsid protein in complex of inhibitor.
Proc Natl Acad Sci U S A 117(30):17992-18001 (2020)
Xia H, Xie X, Zou J, Noble CG, Russell WK, Holthauzen LMF, Choi KH, White MA, Shi PY
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
UniProt ID: P11717 (36-761) Cation-independent mannose-6-phosphate receptor
|
|
|
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Oct 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
UniProt ID: P11717 (36-761) Cation-independent mannose-6-phosphate receptor
|
|
|
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
147 |
nm3 |
|
|
UniProt ID: P11717 (36-761) Cation-independent mannose-6-phosphate receptor
|
|
|
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
UniProt ID: P50897 (28-306) Palmitoyl-protein thioesterase 1
|
|
|
|
| Sample: |
Palmitoyl-protein thioesterase 1 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
UniProt ID: P11717 (36-761) Cation-independent mannose-6-phosphate receptor
UniProt ID: P50897 (28-306) Palmitoyl-protein thioesterase 1
|
|
|
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 81 kDa Homo sapiens protein
Palmitoyl-protein thioesterase 1 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM imidazole, 150 mM NaCl, 5 mM beta glycerol phosphate, 10 mM MnCl2, pH: 6.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 12
|
Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol 3(1):498 (2020)
Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM
|
| RgGuinier |
4.9 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
258 |
nm3 |
|
|
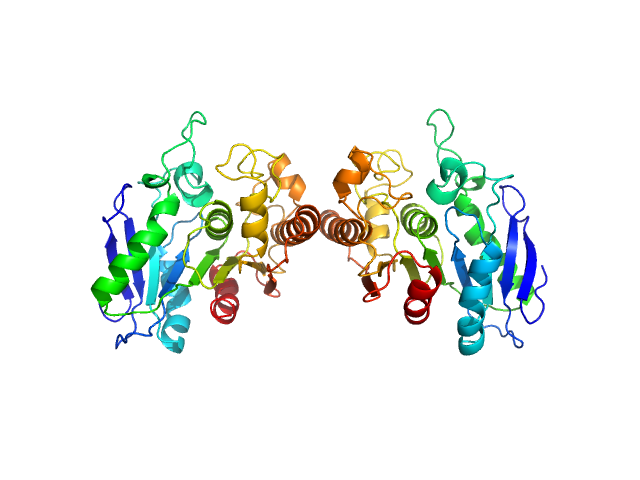
UniProt ID: Q7WSC1 (41-314) Poly(Aspartic acid) hydrolase-1
|
|
|
|
| Sample: |
Poly(Aspartic acid) hydrolase-1 dimer, 65 kDa Sphingomonas sp. KT-1 protein
|
| Buffer: |
20 mM Tris pH 7.0, 100 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 27
|
Structural Characterization of Sphingomonas
sp. KT-1 PahZ1-Catalyzed Biodegradation of Thermally Synthesized Poly(aspartic acid)
ACS Sustainable Chemistry & Engineering (2020)
Brambley C, Bolay A, Salvo H, Jansch A, Yared T, Miller J, Wallen J, Weiland M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
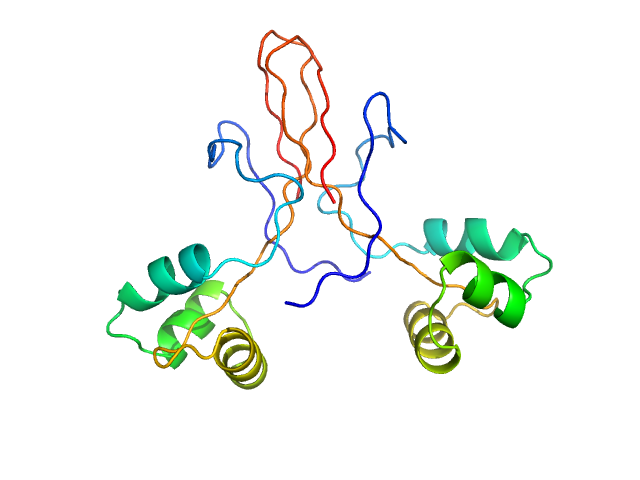
UniProt ID: P03771 (1-68) Protein ninH
|
|
|
|
| Sample: |
Protein ninH dimer, 20 kDa Escherichia phage lambda protein
|
| Buffer: |
150 mM NaCl, 50 mM phosphate buffer (pH 7.4), 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 1
|
A bacteriophage mimic of the bacterial nucleoid-associated protein Fis.
Biochem J (2020)
Chakraborti S, Balakrishnan D, Trotter AJ, Gittens WH, Yang AWH, Jolma A, Paterson JR, Świątek S, Plewka J, Curtis FA, Bowers LY, Pålsson LO, Hughes TR, Taube M, Kozak M, Heddle JG, Sharples GJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
33 |
nm3 |
|
|