UniProt ID: Q9IK91 (1-709) Phosphoprotein
|
|
|
|
| Sample: |
Phosphoprotein tetramer, 317 kDa Nipah henipavirus protein
|
| Buffer: |
20 mM Tris-HCL, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2011 Nov 3
|
Structural Description of the Nipah Virus Phosphoprotein and Its Interaction with STAT1.
Biophys J (2020)
Jensen MR, Yabukarski F, Communie G, Condamine E, Mas C, Volchkova V, Tarbouriech N, Bourhis JM, Volchkov V, Blackledge M, Jamin M
|
| RgGuinier |
10.9 |
nm |
| Dmax |
39.3 |
nm |
|
|
UniProt ID: Q9BQ95 (249-431) Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial
|
|
|

|
| Sample: |
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial dimer, 44 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 7
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
UniProt ID: Q9Y375 (25-327) Complex I intermediate-associated protein 30, mitochondrial
|
|
|
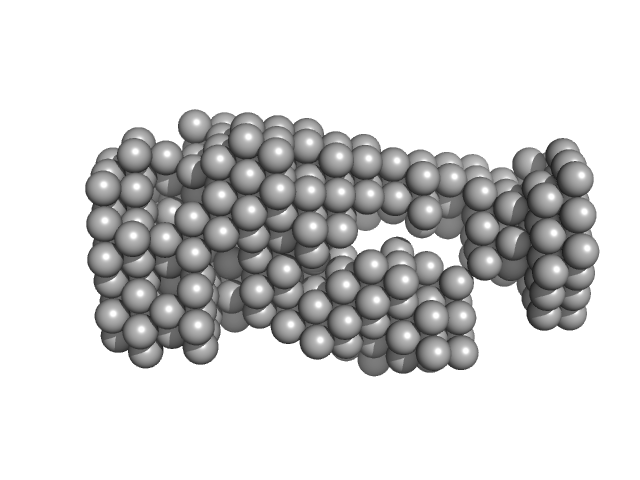
|
| Sample: |
Complex I intermediate-associated protein 30, mitochondrial monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 29
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
UniProt ID: Q9H845 (38-621) Complex I assembly factor ACAD9, mitochondrial
|
|
|
|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial dimer, 132 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 28
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
UniProt ID: P49748 (72-655) Very long-chain specific acyl-CoA dehydrogenase, mitochondrial
|
|
|

|
| Sample: |
Very long-chain specific acyl-CoA dehydrogenase, mitochondrial dimer, 126 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 2
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
215 |
nm3 |
|
|
UniProt ID: Q9H845 (38-621) Complex I assembly factor ACAD9, mitochondrial
UniProt ID: Q9BQ95 (249-431) Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial
|
|
|

|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial dimer, 132 kDa Homo sapiens protein
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial tetramer, 88 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 4
|
Assembly of the mitochondrial Complex I assembly complex suggests a regulatory role for deflavination.
Angew Chem Int Ed Engl (2020)
Giachin G, Jessop M, Bouverot R, Acajjaoui S, Saidi M, Chretien A, Bacia-Verloop M, Signor L, Mas PJ, Favier A, Borel Meneroud E, Hons M, Hart DJ, Kandiah E, Boeri Erba E, Buisson A, Leonard G, Gutsche I, Soler-Lopez M
|
| RgGuinier |
6.0 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
683 |
nm3 |
|
|
UniProt ID: F2R7Z6 (1-401) Alkaline phosphodiesterase I or Nucleotide pyrophosphatase
|
|
|
|
| Sample: |
Alkaline phosphodiesterase I or Nucleotide pyrophosphatase monomer, 46 kDa Streptomyces venezuelae protein
|
| Buffer: |
20 mM HEPES, 200mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 17
|
c-di-AMP hydrolysis by a novel type of phosphodiesterase promotes differentiation of multicellular bacteria
(2019)
Latoscha A, Drexler D, Al-Bassam M, Kaever V, Findlay K, Witte G, Tschowri N
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
UniProt ID: F2R9B6 (6-161) Cyclic di-AMP binding protein (Putative regulatory, ligand-binding protein)
|
|
|
|
| Sample: |
Cyclic di-AMP binding protein (Putative regulatory, ligand-binding protein) dimer, 38 kDa Streptomyces venezuelae protein
|
| Buffer: |
200 mM NaCl, 30 mM NaPi, 5% (v/v) glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 4
|
c-di-AMP hydrolysis by a novel type of phosphodiesterase promotes differentiation of multicellular bacteria
(2019)
Latoscha A, Drexler D, Al-Bassam M, Kaever V, Findlay K, Witte G, Tschowri N
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
UniProt ID: Q13547 (1-482) Histone deacetylase 1
UniProt ID: O60341 (1-852) Lysine-specific histone demethylase 1A
UniProt ID: Q9UKL0 (86-485) REST corepressor 1
|
|
|
|
| Sample: |
Histone deacetylase 1 monomer, 55 kDa Homo sapiens protein
Lysine-specific histone demethylase 1A monomer, 93 kDa Homo sapiens protein
REST corepressor 1 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris/Cl, 50 mM potassium acetate and 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Jan 23
|
Mechanism of Crosstalk between the LSD1 Demethylase and HDAC1 Deacetylase in the CoREST Complex.
Cell Rep 30(8):2699-2711.e8 (2020)
Song Y, Dagil L, Fairall L, Robertson N, Wu M, Ragan TJ, Savva CG, Saleh A, Morone N, Kunze MBA, Jamieson AG, Cole PA, Hansen DF, Schwabe JWR
|
| RgGuinier |
6.0 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
437 |
nm3 |
|
|
UniProt ID: Q15366 (1-365) Poly(rC)-binding protein 2
|
|
|
|
| Sample: |
Poly(rC)-binding protein 2 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jul 16
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
76 |
nm3 |
|
|