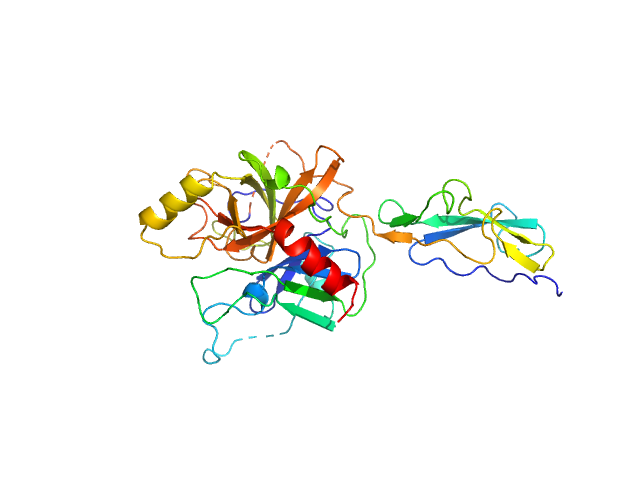
UniProt ID: P00736 (375-705) Complement C1r subcomponent
|
|
|
|
| Sample: |
Complement C1r subcomponent monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 140 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 24
|
A Structural Basis for Inhibition of the Complement Initiator Protease C1r by Lyme Disease Spirochetes.
J Immunol 207(11):2856-2867 (2021)
Garrigues RJ, Powell-Pierce AD, Hammel M, Skare JT, Garcia BL
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
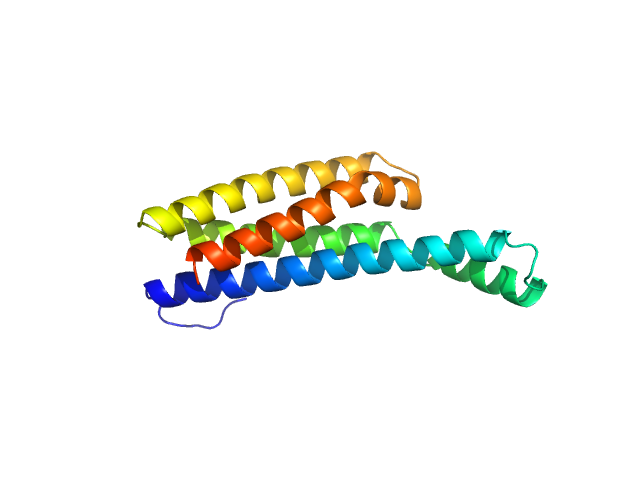
UniProt ID: O50835 (206-348) Fibronectin-binding protein BBK32
|
|
|
|
| Sample: |
Fibronectin-binding protein BBK32 monomer, 17 kDa Borreliella burgdorferi B31 protein
|
| Buffer: |
10 mM HEPES, 140 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 24
|
A Structural Basis for Inhibition of the Complement Initiator Protease C1r by Lyme Disease Spirochetes.
J Immunol 207(11):2856-2867 (2021)
Garrigues RJ, Powell-Pierce AD, Hammel M, Skare JT, Garcia BL
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
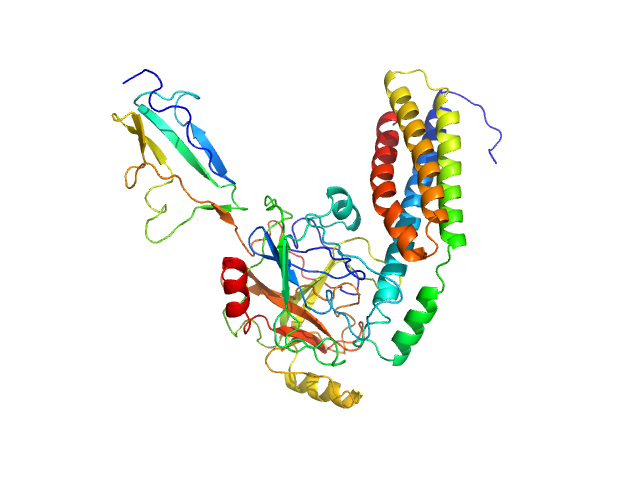
UniProt ID: O50835 (206-348) Fibronectin-binding protein BBK32
UniProt ID: P00736 (376-705) Complement C1r subcomponent
|
|
|
|
| Sample: |
Fibronectin-binding protein BBK32 monomer, 17 kDa Borrelia burgdorferi (strain … protein
Complement C1r subcomponent monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 140 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 24
|
A Structural Basis for Inhibition of the Complement Initiator Protease C1r by Lyme Disease Spirochetes.
J Immunol 207(11):2856-2867 (2021)
Garrigues RJ, Powell-Pierce AD, Hammel M, Skare JT, Garcia BL
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
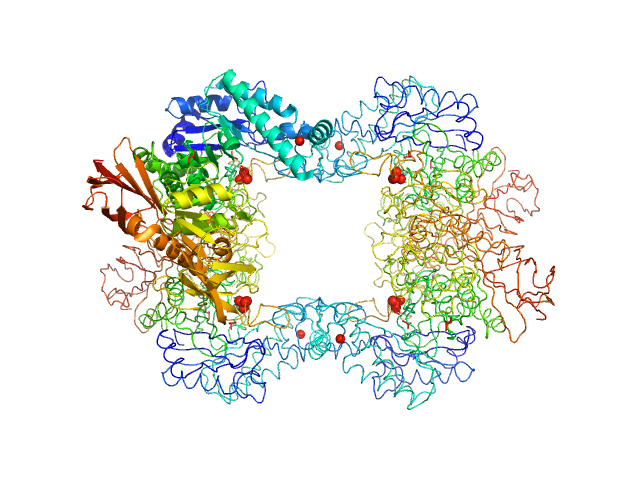
UniProt ID: K0JCW6 (1-660) Alpha-L-fucosidase
|
|
|
|
| Sample: |
Alpha-L-fucosidase tetramer, 297 kDa Paenibacillus thiaminolyticus (Bacillus … protein
|
| Buffer: |
50 mM potassium phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2019 Jul 16
|
The first structure–function study of GH151 α‐ l
‐fucosidase uncovers new oligomerization pattern, active site complementation, and selective substrate specificity
The FEBS Journal (2022)
Koval'ová T, Kovaľ T, Stránský J, Kolenko P, Dušková J, Švecová L, Vodičková P, Spiwok V, Benešová E, Lipovová P, Dohnálek J
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
418 |
nm3 |
|
|
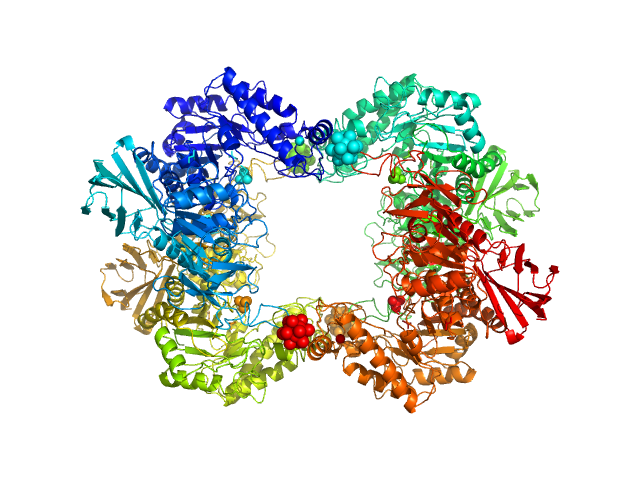
UniProt ID: K0JCW6 (1-660) Alpha-L-fucosidase H503A
|
|
|
|
| Sample: |
Alpha-L-fucosidase H503A tetramer, 297 kDa Paenibacillus thiaminolyticus (Bacillus … protein
|
| Buffer: |
50 mM potassium phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2019 Jul 16
|
The first structure–function study of GH151 α‐ l
‐fucosidase uncovers new oligomerization pattern, active site complementation, and selective substrate specificity
The FEBS Journal (2022)
Koval'ová T, Kovaľ T, Stránský J, Kolenko P, Dušková J, Švecová L, Vodičková P, Spiwok V, Benešová E, Lipovová P, Dohnálek J
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
433 |
nm3 |
|
|
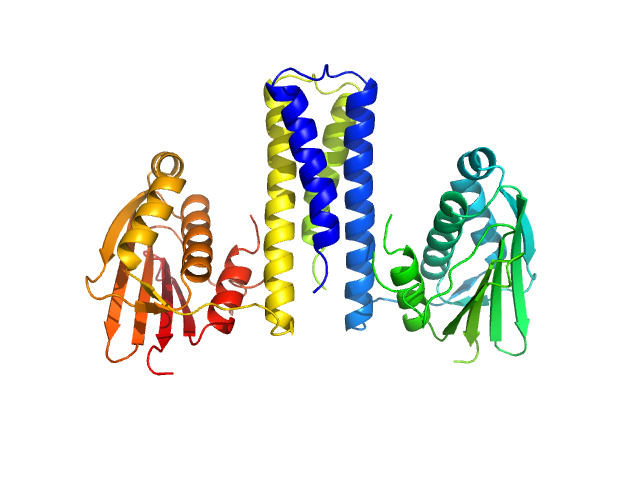
UniProt ID: Q93E22 (130-357) Histidine kinase
|
|
|
|
| Sample: |
Histidine kinase dimer, 51 kDa Acinetobacter baumannii protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Apr 21
|
Proteolysis and multimerization regulate signaling along the two-component regulatory system AdeRS.
iScience 24(5):102476 (2021)
Ouyang Z, Zheng F, Zhu L, Felix J, Wu D, Wu K, Gutsche I, Wu Y, Hwang PM, She J, Wen Y
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
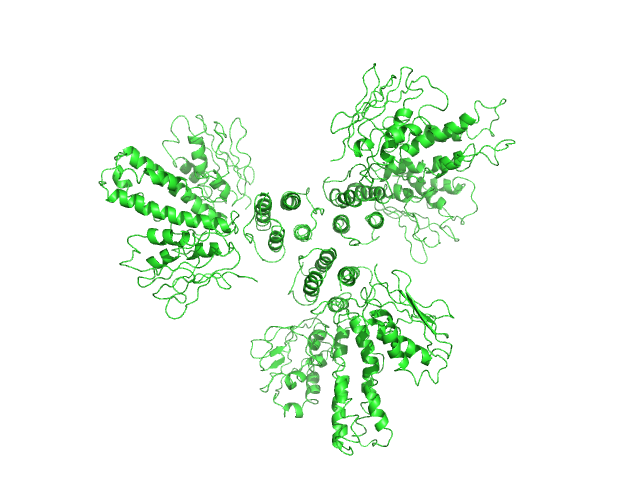
UniProt ID: Q93E22 (84-357) Histidine kinase
|
|
|
|
| Sample: |
Histidine kinase hexamer, 186 kDa Acinetobacter baumannii protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2019 Jun 19
|
Proteolysis and multimerization regulate signaling along the two-component regulatory system AdeRS.
iScience 24(5):102476 (2021)
Ouyang Z, Zheng F, Zhu L, Felix J, Wu D, Wu K, Gutsche I, Wu Y, Hwang PM, She J, Wen Y
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
378 |
nm3 |
|
|
UniProt ID: Q15596-1 (624-773) Nuclear receptor coactivator 2
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDK68_fit1_model1.png)
|
| Sample: |
Nuclear receptor coactivator 2 monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 50 mM BisTris pH 6.8, and 0.5 mM EDTA, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 14
|
Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J Mol Biol 433(9):166899 (2021)
Senicourt L, le Maire A, Allemand F, Carvalho JE, Guee L, Germain P, Schubert M, Bernadó P, Bourguet W, Sibille N
|
| RgGuinier |
3.7 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
UniProt ID: Q14258 (189-630) E3 ubiquitin/ISG15 ligase TRIM25
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 19
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
6.8 |
nm |
| Dmax |
30.2 |
nm |
|
|
UniProt ID: Q14258 (189-630) E3 ubiquitin/ISG15 ligase TRIM25
UniProt ID: None (None-None) pre-let-7-a-1@1
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
Pre-let-7-a-1@1 monomer, 9 kDa synthetic construct RNA
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 14
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.8 |
nm |
|
|