UniProt ID: Q9VV74 (186-220) Survival motor neuron protein
UniProt ID: P0AEX9 (1-396) Maltose/maltodextrin-binding periplasmic protein
|
|
|
|
| Sample: |
Survival motor neuron protein tetramer, 15 kDa Drosophila melanogaster protein
Maltose/maltodextrin-binding periplasmic protein tetramer, 174 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM Na/KPO4 pH 7.0, 300 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 19
|
Assembly of higher-order SMN oligomers is essential for metazoan viability and requires an exposed structural motif present in the YG zipper dimer.
Nucleic Acids Res 49(13):7644-7664 (2021)
Gupta K, Wen Y, Ninan NS, Raimer AC, Sharp R, Spring AM, Sarachan KL, Johnson MC, Van Duyne GD, Matera AG
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
340 |
nm3 |
|
|
UniProt ID: P0AEX9 (1-396) Maltose/maltodextrin-binding periplasmic protein
UniProt ID: Q9VV74 (186-220) Survival motor neuron protein
|
|
|
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein octamer, 347 kDa Escherichia coli (strain … protein
Survival motor neuron protein octamer, 31 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Na/KPO4 pH 7.0, 300 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 19
|
Assembly of higher-order SMN oligomers is essential for metazoan viability and requires an exposed structural motif present in the YG zipper dimer.
Nucleic Acids Res 49(13):7644-7664 (2021)
Gupta K, Wen Y, Ninan NS, Raimer AC, Sharp R, Spring AM, Sarachan KL, Johnson MC, Van Duyne GD, Matera AG
|
| RgGuinier |
5.6 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
800 |
nm3 |
|
|
UniProt ID: Q9VVX0 (1-245) Protein Gemin2
UniProt ID: Q9VV74 (1-226) Survival motor neuron protein
|
|
|
|
| Sample: |
Protein Gemin2 octamer, 230 kDa Drosophila melanogaster protein
Survival motor neuron protein octamer, 197 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Na/KPO4 pH 7.0, 300 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 19
|
Assembly of higher-order SMN oligomers is essential for metazoan viability and requires an exposed structural motif present in the YG zipper dimer.
Nucleic Acids Res 49(13):7644-7664 (2021)
Gupta K, Wen Y, Ninan NS, Raimer AC, Sharp R, Spring AM, Sarachan KL, Johnson MC, Van Duyne GD, Matera AG
|
| RgGuinier |
7.8 |
nm |
| Dmax |
28.7 |
nm |
| VolumePorod |
2660 |
nm3 |
|
|
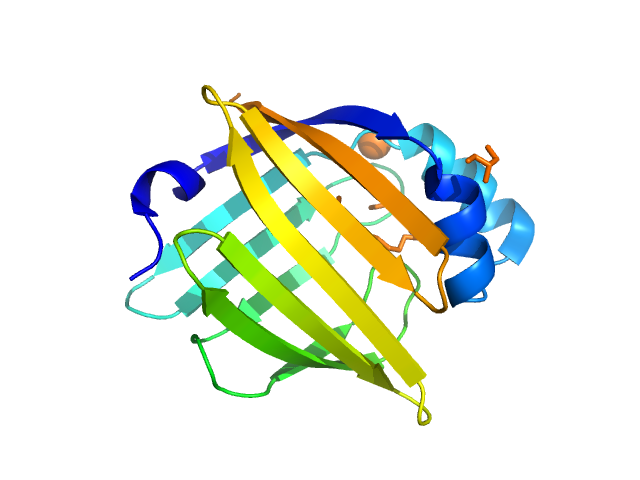
UniProt ID: P02689 (1-132) Myelin P2 protein
|
|
|
|
| Sample: |
Myelin P2 protein monomer, 15 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jul 22
|
Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
FEBS J (2021)
Uusitalo M, Klenow MB, Laulumaa S, Blakeley MP, Simonsen AC, Ruskamo S, Kursula P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.2 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
UniProt ID: P51449 (25-518) Retinoid-related orphan receptor-gamma
UniProt ID: None (None-None) Classic-RORgamma Response Element
|
|
|
|
| Sample: |
Retinoid-related orphan receptor-gamma monomer, 56 kDa Homo sapiens protein
Classic-RORgamma Response Element dimer, 19 kDa Homo sapiens DNA
|
| Buffer: |
25 mM HEPES, 150 mM TCEP, 2% Glycerol, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Nov 20
|
Conformational Changes of RORγ During Response Element Recognition and Coregulator Engagement
Journal of Molecular Biology :167258 (2021)
Strutzenberg T, Zhu Y, Novick S, Garcia-Ordonez R, Doebelin C, He Y, Ra Chang M, Kamenecka T, Edwards D, Griffin P
|
| RgGuinier |
5.5 |
nm |
| Dmax |
22.9 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
UniProt ID: P51449 (25-518) Retinoid-related orphan receptor-gamma
UniProt ID: None (None-None) Variant-RORgamma Response Element
|
|
|
|
| Sample: |
Retinoid-related orphan receptor-gamma monomer, 56 kDa Homo sapiens protein
Variant-RORgamma Response Element dimer, 18 kDa Homo sapiens DNA
|
| Buffer: |
25 mM HEPES, 150 mM TCEP, 2% Glycerol, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Nov 20
|
Conformational Changes of RORγ During Response Element Recognition and Coregulator Engagement
Journal of Molecular Biology :167258 (2021)
Strutzenberg T, Zhu Y, Novick S, Garcia-Ordonez R, Doebelin C, He Y, Ra Chang M, Kamenecka T, Edwards D, Griffin P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
22.1 |
nm |
| VolumePorod |
112 |
nm3 |
|
|
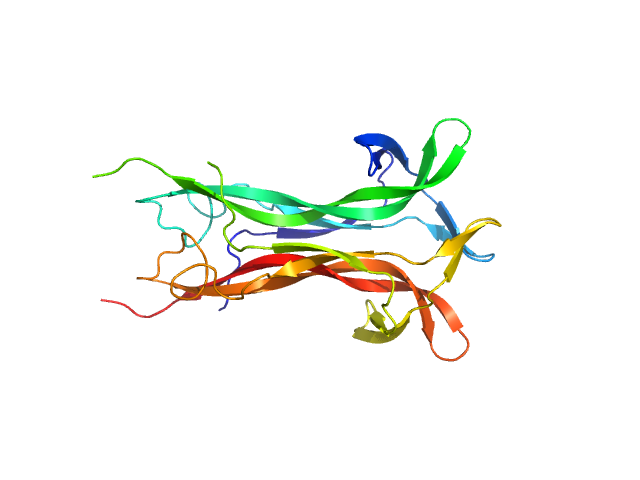
UniProt ID: P01138 (131-239) Beta-nerve growth factor
|
|
|
|
| Sample: |
Beta-nerve growth factor dimer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na-phosphate, 1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Jul 2
|
The conundrum of the high-affinity NGF binding site formation unveiled?
Biophys J 108(3):687-97 (2015)
Covaceuszach S, Konarev PV, Cassetta A, Paoletti F, Svergun DI, Lamba D, Cattaneo A
|
|
|
UniProt ID: P54252 (1-361) Ataxin-3
|
|
|

|
| Sample: |
Ataxin-3 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
6.9 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
425 |
nm3 |
|
|
UniProt ID: Q71U36 (1-451) Tubulin alpha-1A chain
|
|
|
|
| Sample: |
Tubulin alpha-1A chain monomer, 50 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
7.0 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
340 |
nm3 |
|
|
UniProt ID: P54252 (1-361) Ataxin-3
UniProt ID: Q71U36 (1-451) Tubulin alpha-1A chain
|
|
|

|
| Sample: |
Ataxin-3 monomer, 41 kDa Homo sapiens protein
Tubulin alpha-1A chain monomer, 50 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Dec 3
|
Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome.
Int J Biochem Cell Biol 51:58-64 (2014)
Bonanomi M, Mazzucchelli S, D'Urzo A, Nardini M, Konarev PV, Invernizzi G, Svergun DI, Vanoni M, Regonesi ME, Tortora P
|
| RgGuinier |
8.4 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
900 |
nm3 |
|
|