UniProt ID: Q9UK39 (None-None) Nocturnin - Deletion construct - Δ107-120
|
|
|
|
| Sample: |
Nocturnin - Deletion construct - Δ107-120 monomer, 40 kDa protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, 10% glycerol, 5 mM MgCl2, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Feb 7
|
The Disordered Amino Terminus of the Circadian Enzyme Nocturnin Modulates Its NADP(H) Phosphatase Activity by Changing Protein Dynamics.
Biochemistry (2022)
Wickramaratne AC, Li L, Hopkins JB, Joachimiak LA, Green CB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
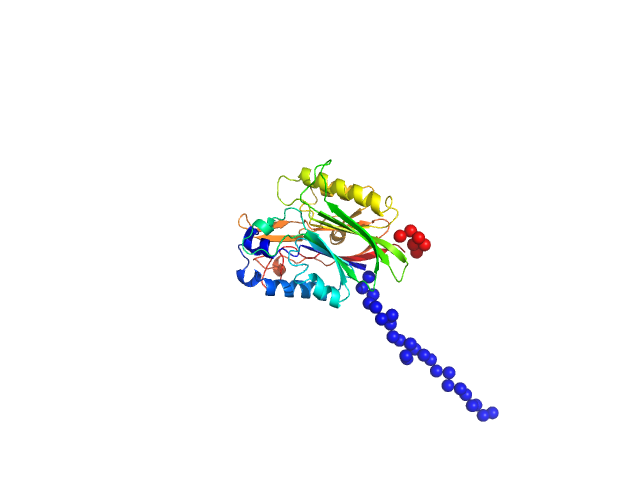
UniProt ID: Q9UK39 (118-431) Nocturnin - N terminus truncated (118-NOCT)
|
|
|
|
| Sample: |
Nocturnin - N terminus truncated (118-NOCT) monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, 10% glycerol, 5 mM MgCl2, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Feb 7
|
The Disordered Amino Terminus of the Circadian Enzyme Nocturnin Modulates Its NADP(H) Phosphatase Activity by Changing Protein Dynamics.
Biochemistry (2022)
Wickramaratne AC, Li L, Hopkins JB, Joachimiak LA, Green CB
|
| RgGuinier |
2.3 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
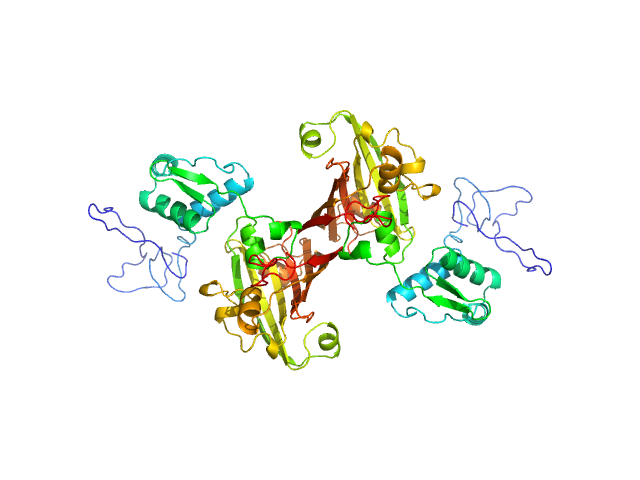
UniProt ID: O86331 (None-None) Recombinant Mycobacterium tuberculosis H37Rv
|
|
|
|
| Sample: |
Recombinant Mycobacterium tuberculosis H37Rv dimer, 65 kDa Mycobacterium tuberculosis H37Rv protein
|
| Buffer: |
25 mM HEPES Buffer; 400 mM NaCl,, pH: 7.2 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 2
|
Structural and Functional Characterization of Rv0792c from Mycobacterium tuberculosis: Identifying Small Molecule Inhibitor against HutC Protein.
Microbiol Spectr :e0197322 (2022)
Chauhan NK, Anand A, Sharma A, Dhiman K, Gosain TP, Singh P, Singh P, Khan E, Chattopadhyay G, Kumar A, Sharma D, Ashish, Sharma TK, Singh R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
|
|
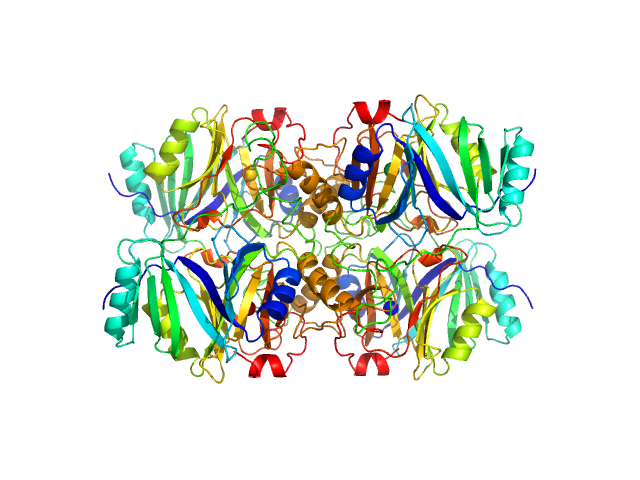
UniProt ID: A0A0N1BQH2 (1-311) Metapyrocatechase
|
|
|
|
| Sample: |
Metapyrocatechase tetramer, 139 kDa Novosphingobium sp. AAP93 protein
|
| Buffer: |
20 mM Tris-HCl 250 mM NaCl, pH 8.0, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Dec 20
|
Shape-function of a novel metapyrocatechase, RW4-MPC: Metagenomics to SAXS data based insight into deciphering regulators of function.
Int J Biol Macromol 188:1012-1024 (2021)
Vasudeva G, Sidhu C, Kalidas N, Ashish, Pinnaka AK
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.2 |
nm |
|
|
UniProt ID: Q06671 (1-453) Autophagy-related protein 23
|
|
|
|
| Sample: |
Autophagy-related protein 23 dimer, 104 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 0.2 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Nov 3
|
Dimerization-dependent membrane tethering by Atg23 is essential for yeast autophagy.
Cell Rep 39(3):110702 (2022)
Hawkins WD, Leary KA, Andhare D, Popelka H, Klionsky DJ, Ragusa MJ
|
| RgGuinier |
7.4 |
nm |
| Dmax |
29.3 |
nm |
| VolumePorod |
378 |
nm3 |
|
|
UniProt ID: Q06671 (1-453) Autophagy-related protein 23 LIL Mutant
|
|
|
|
| Sample: |
Autophagy-related protein 23 LIL Mutant monomer, 52 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 0.2 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 May 8
|
Dimerization-dependent membrane tethering by Atg23 is essential for yeast autophagy.
Cell Rep 39(3):110702 (2022)
Hawkins WD, Leary KA, Andhare D, Popelka H, Klionsky DJ, Ragusa MJ
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
UniProt ID: Q06671 (1-453) Autophagy-related protein 23 RKK Mutant
|
|
|
|
| Sample: |
Autophagy-related protein 23 RKK Mutant dimer, 104 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 0.2 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 May 8
|
Dimerization-dependent membrane tethering by Atg23 is essential for yeast autophagy.
Cell Rep 39(3):110702 (2022)
Hawkins WD, Leary KA, Andhare D, Popelka H, Klionsky DJ, Ragusa MJ
|
| RgGuinier |
7.6 |
nm |
| Dmax |
34.5 |
nm |
| VolumePorod |
365 |
nm3 |
|
|
UniProt ID: P18827 (23-254) Syndecan-1
|
|
|
|
| Sample: |
Syndecan-1 monomer, 27 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 26
|
Extended disorder at the cell surface: the conformational landscape of the ectodomains of syndecans
Matrix Biology Plus :100081 (2021)
Gondelaud F, Bouakil M, Le Fèvre A, Erica Miele A, Chirot F, Duclos B, Liwo A, Ricard-Blum S
|
|
|
UniProt ID: P34741 (19-144) Syndecan-2
|
|
|
|
| Sample: |
Syndecan-2 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 31
|
Extended disorder at the cell surface: the conformational landscape of the ectodomains of syndecans
Matrix Biology Plus :100081 (2021)
Gondelaud F, Bouakil M, Le Fèvre A, Erica Miele A, Chirot F, Duclos B, Liwo A, Ricard-Blum S
|
|
|
UniProt ID: O75056 (47-387) Syndecan-3
|
|
|
|
| Sample: |
Syndecan-3 monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 May 29
|
Extended disorder at the cell surface: the conformational landscape of the ectodomains of syndecans
Matrix Biology Plus :100081 (2021)
Gondelaud F, Bouakil M, Le Fèvre A, Erica Miele A, Chirot F, Duclos B, Liwo A, Ricard-Blum S
|
|
|