UniProt ID: Q8BX22 (870-940) Sal-like protein 4
|
|
|
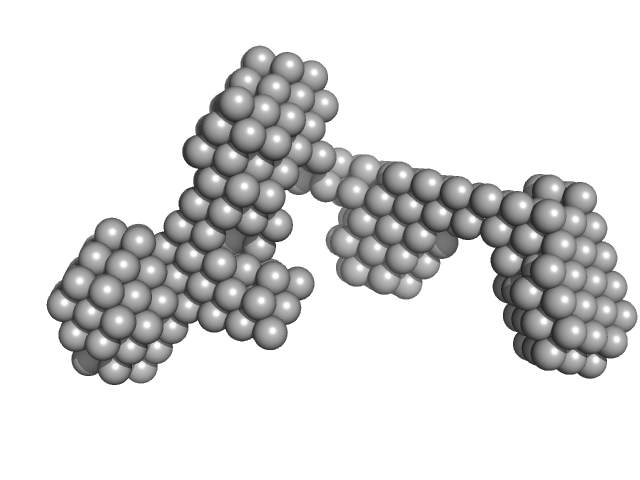
|
| Sample: |
Sal-like protein 4 monomer, 8 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, pH 7.5, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
Structure of SALL4 zinc finger domain reveals link between AT-rich DNA binding and Okihiro syndrome.
Life Sci Alliance 6(3) (2023)
Watson JA, Pantier R, Jayachandran U, Chhatbar K, Alexander-Howden B, Kruusvee V, Prendecki M, Bird A, Cook AG
|
| RgGuinier |
2.1 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
11 |
nm3 |
|
|
UniProt ID: Q8BX22 (870-940) Sal-like protein 4
UniProt ID: None (None-None) AT-rich dsDNA
|
|
|
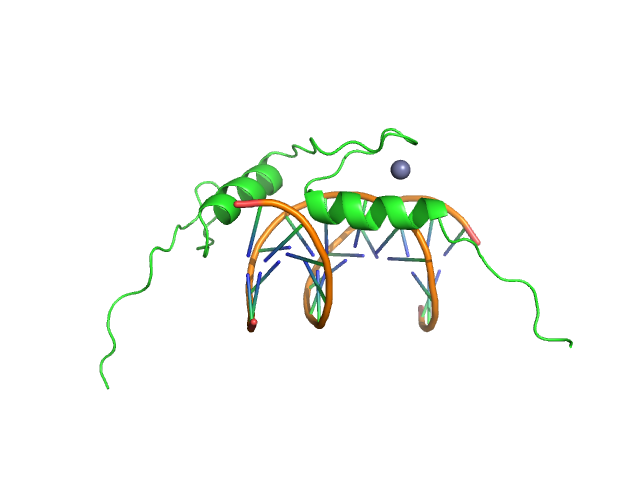
|
| Sample: |
Sal-like protein 4 monomer, 8 kDa Mus musculus protein
AT-rich dsDNA monomer, 7 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, pH 7.5, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
Structure of SALL4 zinc finger domain reveals link between AT-rich DNA binding and Okihiro syndrome.
Life Sci Alliance 6(3) (2023)
Watson JA, Pantier R, Jayachandran U, Chhatbar K, Alexander-Howden B, Kruusvee V, Prendecki M, Bird A, Cook AG
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.1 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
UniProt ID: Q132Y4 (1-481) TIR domain-containing protein
UniProt ID: Q132Y5 (1-507) Uncharacterized protein (PIWI)
|
|
|
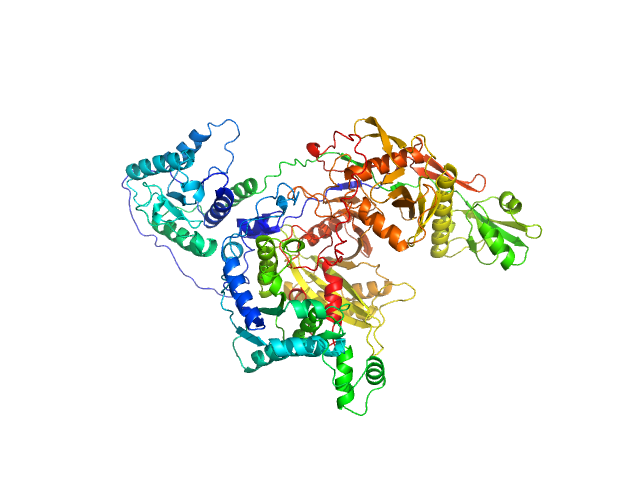
|
| Sample: |
TIR domain-containing protein monomer, 57 kDa Rhodopseudomonas palustris (strain … protein
Uncharacterized protein (PIWI) monomer, 56 kDa Rhodopseudomonas palustris (strain … protein
|
| Buffer: |
20 mM Tris-HCl, pH8.0, 200 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 27
|
Complex of Rhodopseudomonas palustris pAgo with TIR-like effector protein
Elena Manakova
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
192 |
nm3 |
|
|
UniProt ID: P02763 (19-183) Alpha-1-acid glycoprotein 1
|
|
|
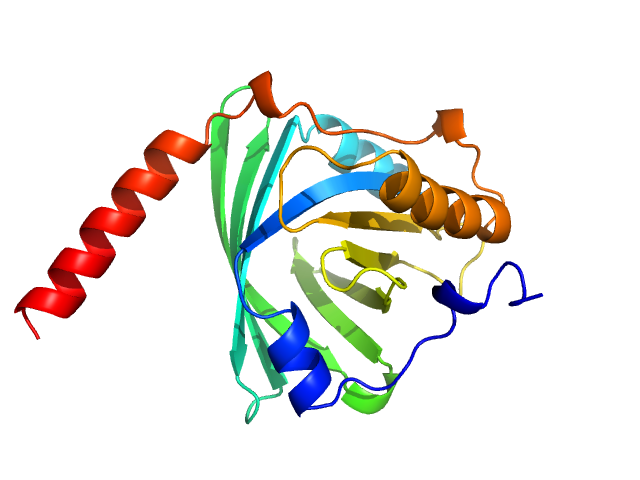
|
| Sample: |
Alpha-1-acid glycoprotein 1 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2021 Jan 12
|
SAXS data based glycosylated models of human alpha-1-acid glycorprotein, a key player in health, disease and drug circulation.
J Biomol Struct Dyn :1-15 (2025)
Kalidas N, Peddada N, Pandey K, Ashish
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
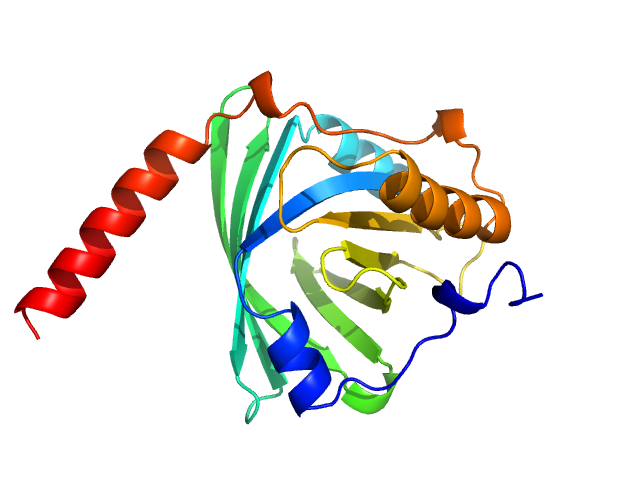
UniProt ID: P02763 (19-183) Alpha-1-acid glycoprotein 1
|
|
|
|
| Sample: |
Alpha-1-acid glycoprotein 1 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2021 Jan 12
|
SAXS data based glycosylated models of human alpha-1-acid glycorprotein, a key player in health, disease and drug circulation.
J Biomol Struct Dyn :1-15 (2025)
Kalidas N, Peddada N, Pandey K, Ashish
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
UniProt ID: P9WPI1 (1-278) ESX-5 secretion system protein EccA5
|
|
|

|
| Sample: |
ESX-5 secretion system protein EccA5 monomer, 31 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 9
|
Structural and Biophysical characterization of N-terminal domain of EccA5 from Mycobacterium tuberculosis.
Jyoti Vishwakarma
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
UniProt ID: Q7AL96 (None-None) DNA-binding protein
|
|
|
|
| Sample: |
DNA-binding protein monomer, 13 kDa Mesorhizobium loti protein
|
| Buffer: |
150 mM Tris-HCl, 300 mM NaCl, 5% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 19
|
Crystallographic and X-ray scattering study of RdfS, a recombination directionality factor from an integrative and conjugative element
Acta Crystallographica Section D Structural Biology 78(10) (2022)
Verdonk C, Marshall A, Ramsay J, Bond C
|
| RgGuinier |
2.2 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
UniProt ID: Q02817 (4356-5150) Human Mucin 2 C-terminal
|
|
|
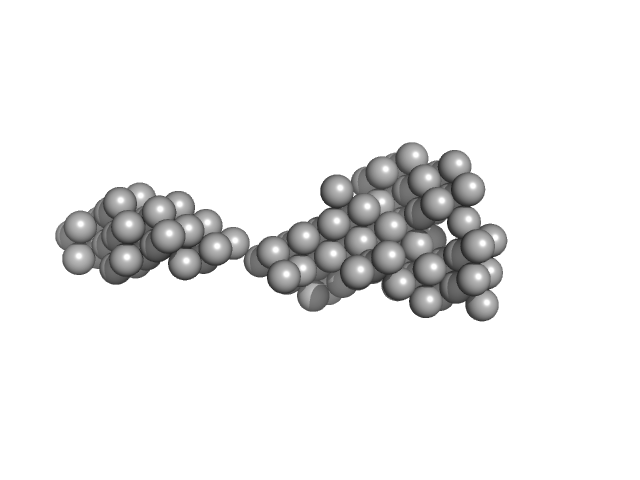
|
| Sample: |
Human Mucin 2 C-terminal dimer, 240 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 10 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 20
|
The intestinal MUC2 mucin C-terminus is stabilized by an extra disulfide bond in comparison to von Willebrand factor and other gel-forming mucins
Nature Communications 14(1) (2023)
Gallego P, Garcia-Bonete M, Trillo-Muyo S, Recktenwald C, Johansson M, Hansson G
|
| RgGuinier |
8.4 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
684 |
nm3 |
|
|
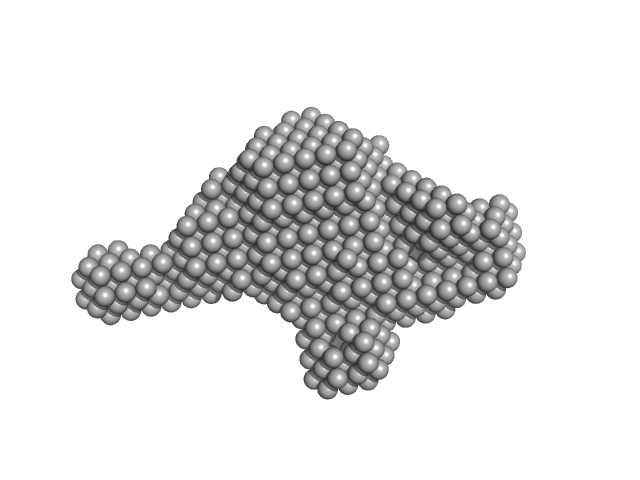
UniProt ID: P20936 (174-1047) Ras GTPase-activating protein 1
|
|
|
|
| Sample: |
Ras GTPase-activating protein 1 monomer, 101 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8 350 mM NaCl 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Dec 12
|
Tandem engagement of phosphotyrosines by the dual SH2 domains of p120RasGAP.
Structure (2022)
Stiegler AL, Vish KJ, Boggon TJ
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
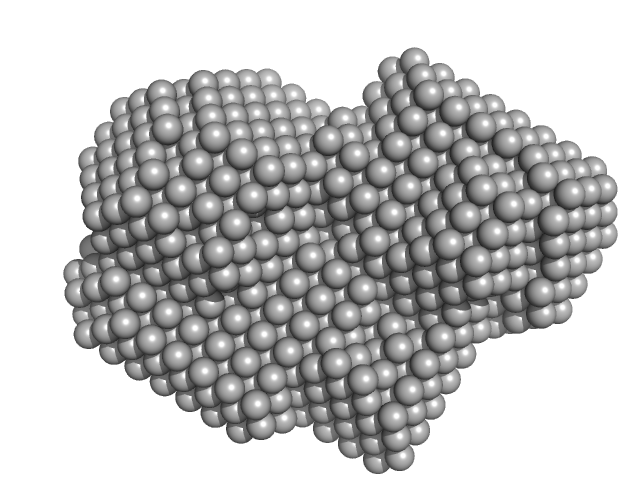
UniProt ID: P20936 (174-1047) Ras GTPase-activating protein 1
UniProt ID: Q9NRY4 (1083-1111) Rho GTPase-activating protein 35
|
|
|
|
| Sample: |
Ras GTPase-activating protein 1 monomer, 101 kDa Homo sapiens protein
Rho GTPase-activating protein 35 monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8 350 mM NaCl 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Dec 11
|
Tandem engagement of phosphotyrosines by the dual SH2 domains of p120RasGAP.
Structure (2022)
Stiegler AL, Vish KJ, Boggon TJ
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
41 |
nm3 |
|
|