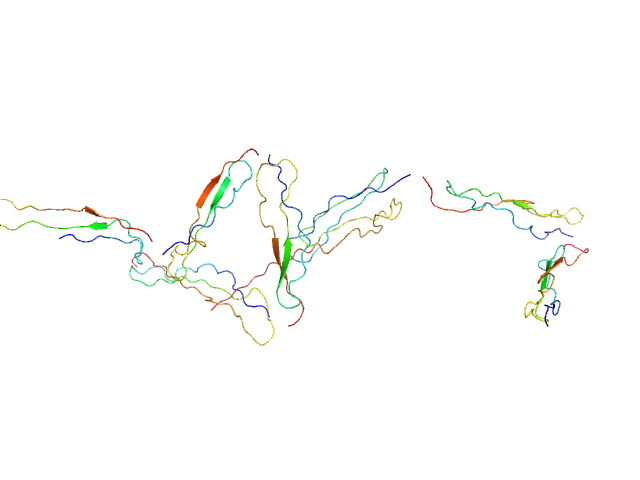
UniProt ID: P27918 (None-None) Human recombinant Properdin TSR 0-3
UniProt ID: P27918 (None-None) Human recombinant Properdin TSR 4-6
|
|
|
|
| Sample: |
Human recombinant Properdin TSR 0-3 monomer, 25 kDa Homo sapiens protein
Human recombinant Properdin TSR 4-6 monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Aug 16
|
Functional and structural insight into properdin control of complement alternative pathway amplification.
EMBO J 36(8):1084-1099 (2017)
Pedersen DV, Roumenina L, Jensen RK, Gadeberg TA, Marinozzi C, Picard C, Rybkine T, Thiel S, Sørensen UB, Stover C, Fremeaux-Bacchi V, Andersen GR
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.0 |
nm |
|
|
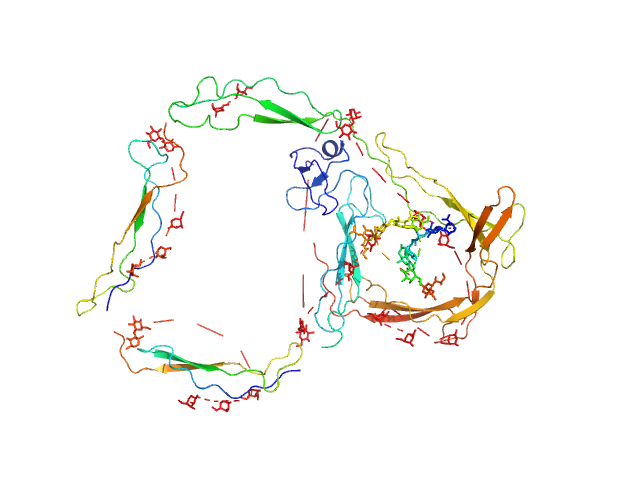
UniProt ID: P27918 (None-None) E244K Human Properdin
|
|
|
|
| Sample: |
E244K Human Properdin monomer, 49 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 27
|
Functional and structural insight into properdin control of complement alternative pathway amplification.
EMBO J 36(8):1084-1099 (2017)
Pedersen DV, Roumenina L, Jensen RK, Gadeberg TA, Marinozzi C, Picard C, Rybkine T, Thiel S, Sørensen UB, Stover C, Fremeaux-Bacchi V, Andersen GR
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
|
|
UniProt ID: Q15554 (42-86) Basic domain of telomeric repeat-binding factor 2
UniProt ID: (None-None) telomere DNA duplex
|
|
|
|
| Sample: |
Basic domain of telomeric repeat-binding factor 2 monomer, 5 kDa Homo sapiens protein
Telomere DNA duplex monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 50 mM LiCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 May 3
|
Basic domain of telomere guardian TRF2 reduces D-loop unwinding whereas Rap1 restores it.
Nucleic Acids Res 45(21):12170-12180 (2017)
Necasová I, Janoušková E, Klumpler T, Hofr C
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
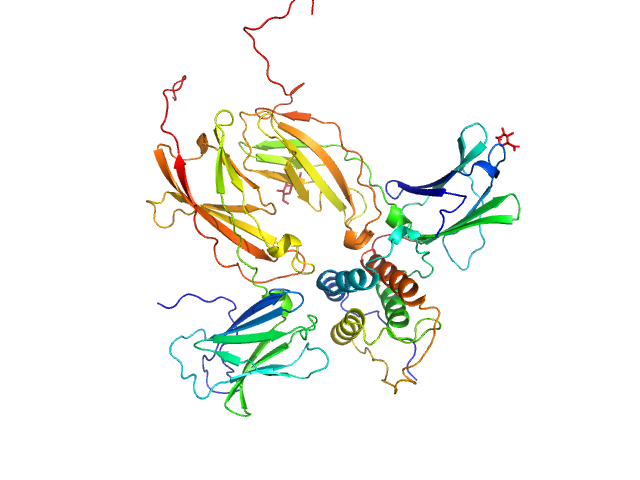
UniProt ID: Q969D9 (None-None) Thymic stromal lymphopoietin
UniProt ID: P16871 (21-239) Interleukin-7 receptor subunit alpha
UniProt ID: Q9HC73 (25-221) Cytokine receptor-like factor 2
|
|
|
|
| Sample: |
Thymic stromal lymphopoietin monomer, 15 kDa Homo sapiens protein
Interleukin-7 receptor subunit alpha monomer, 26 kDa Homo sapiens protein
Cytokine receptor-like factor 2 monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 8
|
Structure and antagonism of the receptor complex mediated by human TSLP in allergy and asthma.
Nat Commun 8:14937 (2017)
Verstraete K, Peelman F, Braun H, Lopez J, Van Rompaey D, Dansercoer A, Vandenberghe I, Pauwels K, Tavernier J, Lambrecht BN, Hammad H, De Winter H, Beyaert R, Lippens G, Savvides SN
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
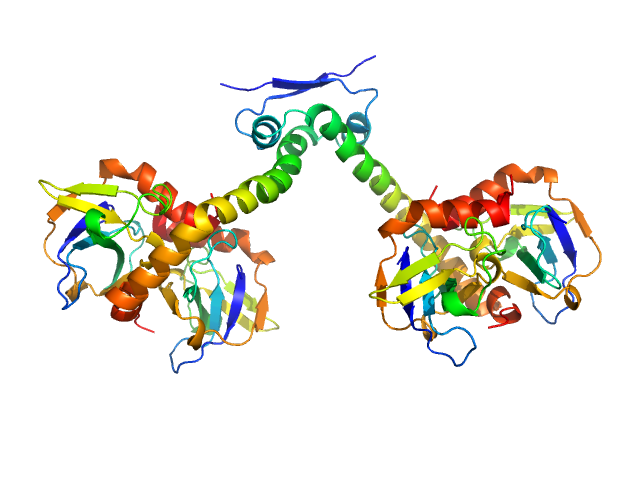
UniProt ID: P62554 (1-101) Toxin CcdB
UniProt ID: P62552 (1-72) Antitoxin CcdA
UniProt ID: P62554 (None-None) Toxin CcdB
|
|
|
|
| Sample: |
Toxin CcdB dimer, 23 kDa Escherichia coli protein
Antitoxin CcdA dimer, 17 kDa Escherichia coli protein
Toxin CcdB dimer, 23 kDa Escherichia coli protein
|
| Buffer: |
10 mM Tris 50 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Jul 24
|
Molecular mechanism governing ratio-dependent transcription regulation in the ccdAB operon.
Nucleic Acids Res 45(6):2937-2950 (2017)
Vandervelde A, Drobnak I, Hadži S, Sterckx YG, Welte T, De Greve H, Charlier D, Efremov R, Loris R, Lah J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
UniProt ID: P42641 (1-390) GTPase ObgE/CgtA
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 uM GDP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
94673 |
nm3 |
|
|
UniProt ID: P42641 (1-340) GTPase ObgE/CgtA
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 uM GDP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, on 2015 Feb 1
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
70673 |
nm3 |
|
|
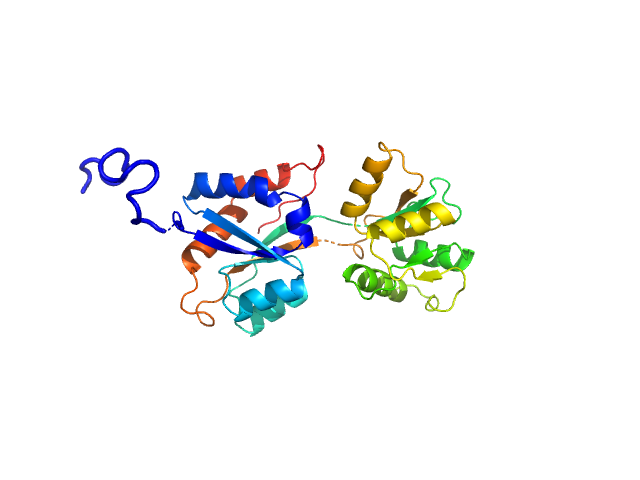
UniProt ID: Q316W1 (None-None) ABC transporter periplasmic substrate-binding protein
|
|
|
|
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 7
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
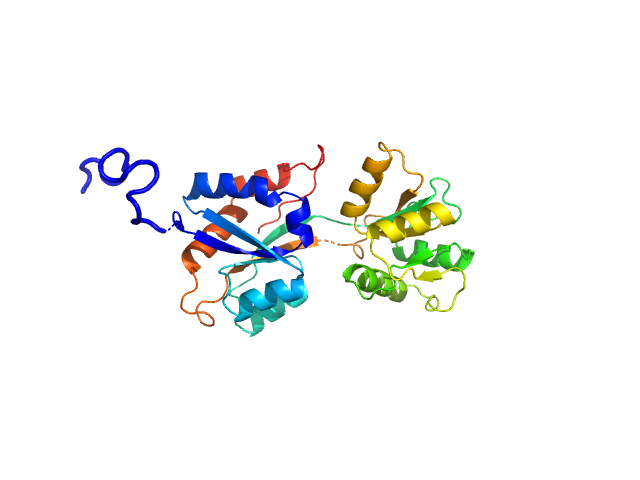
UniProt ID: Q316W1 (None-None) ABC transporter periplasmic substrate-binding protein
|
|
|
|
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jun 21
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
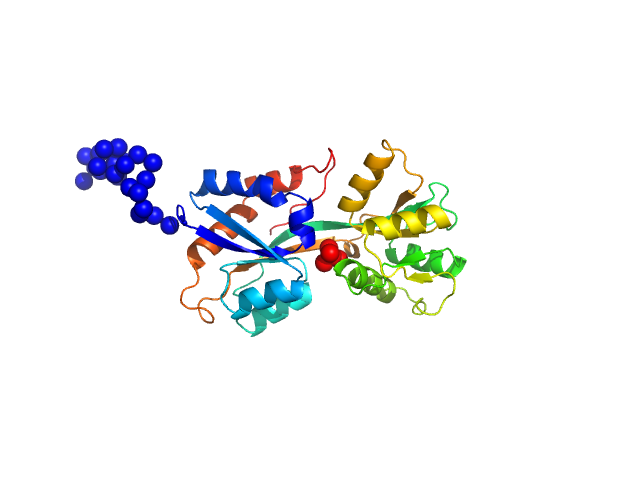
UniProt ID: Q316W1 (None-None) ABC transporter periplasmic substrate-binding protein
|
|
|
|
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 7
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|