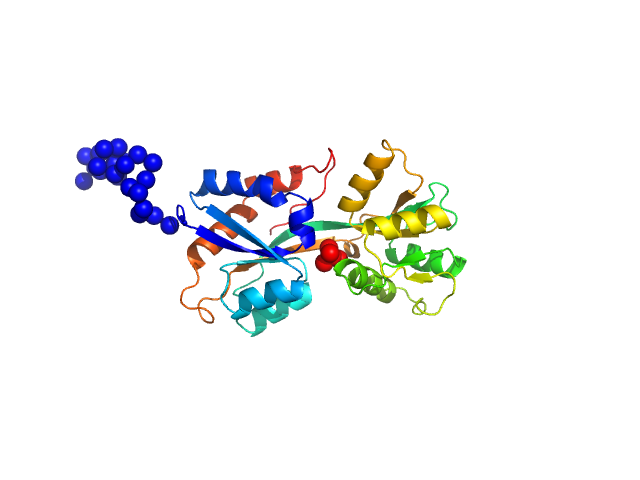
UniProt ID: Q316W1 (None-None) ABC transporter periplasmic substrate-binding protein
|
|
|
|
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jun 21
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
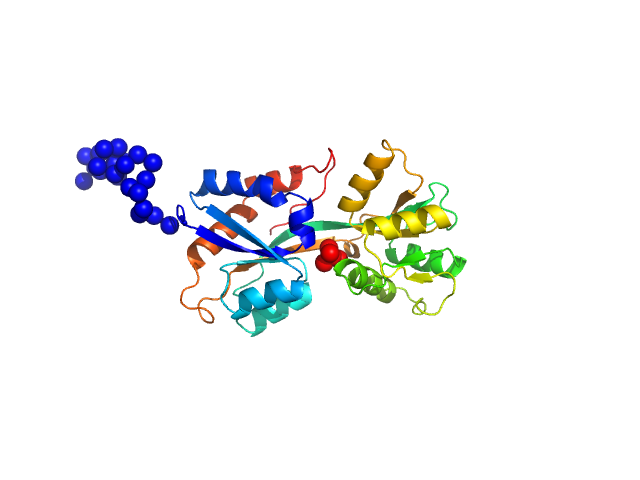
UniProt ID: Q316W1 (None-None) ABC transporter periplasmic substrate-binding protein
|
|
|
|
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 7
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
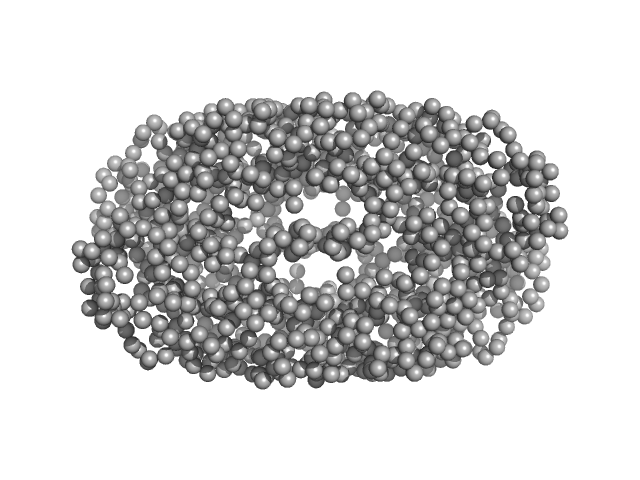
UniProt ID: A0A0K2H6I1 (1-300) Geobacillus stearothermophilus DnaB1-300
|
|
|
|
| Sample: |
Geobacillus stearothermophilus DnaB1-300 tetramer, 138 kDa Geobacillus stearothermophilus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl and 5 mM β-ME, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 Oct 18
|
Structural analyses of the bacterial primosomal protein DnaB reveal that it is a tetramer and forms a complex with a primosomal re-initiation protein.
J Biol Chem 292(38):15744-15757 (2017)
Li YC, Naveen V, Lin MG, Hsiao CD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
315 |
nm3 |
|
|
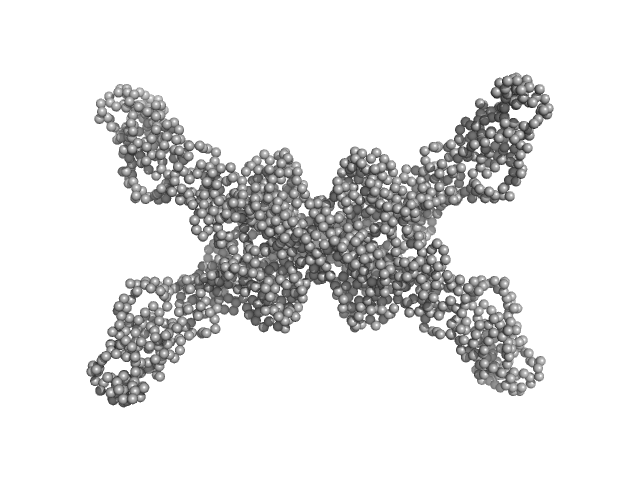
UniProt ID: A0A0K2H6I1 (None-None) Geobacillus stearothermophilus DnaB full-length
|
|
|
|
| Sample: |
Geobacillus stearothermophilus DnaB full-length tetramer, 214 kDa Geobacillus stearothermophilus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl and 5 mM β-ME, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 Mar 12
|
Structural analyses of the bacterial primosomal protein DnaB reveal that it is a tetramer and forms a complex with a primosomal re-initiation protein.
J Biol Chem 292(38):15744-15757 (2017)
Li YC, Naveen V, Lin MG, Hsiao CD
|
| RgGuinier |
5.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
432 |
nm3 |
|
|
UniProt ID: E0TU96 (528-669) Putative DNA binding protein
|
|
|

|
| Sample: |
Putative DNA binding protein dimer, 37 kDa Bacillus subtilis subsp. … protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2016 Aug 17
|
YeeF, a polymorphic toxin from Bacillus subtilis, is a ribosomal RNase with promiscuous DNA binding property
Krishan Gopal
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
UniProt ID: E0TU95 (None-None) YezG, cognate immunity protein of YeeF
|
|
|

|
| Sample: |
YezG, cognate immunity protein of YeeF monomer, 19 kDa Bacillus subtilis subsp. … protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2016 Aug 17
|
YeeF, a polymorphic toxin from Bacillus subtilis, is a ribosomal RNase with promiscuous DNA binding property
Krishan Gopal
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: E0TU96 (None-None) Putative DNA binding protein
UniProt ID: E0TU95 (None-None) YezG, cognate immunity protein of YeeF
|
|
|

|
| Sample: |
Putative DNA binding protein dimer, 37 kDa Bacillus subtilis subsp. … protein
YezG, cognate immunity protein of YeeF monomer, 19 kDa Bacillus subtilis subsp. … protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2016 Aug 18
|
YeeF, a polymorphic toxin from Bacillus subtilis, is a ribosomal RNase with promiscuous DNA binding property
Krishan Gopal
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
UniProt ID: Q16543 (None-None) Hsp90 co-chaperone Cdc37
|
|
|
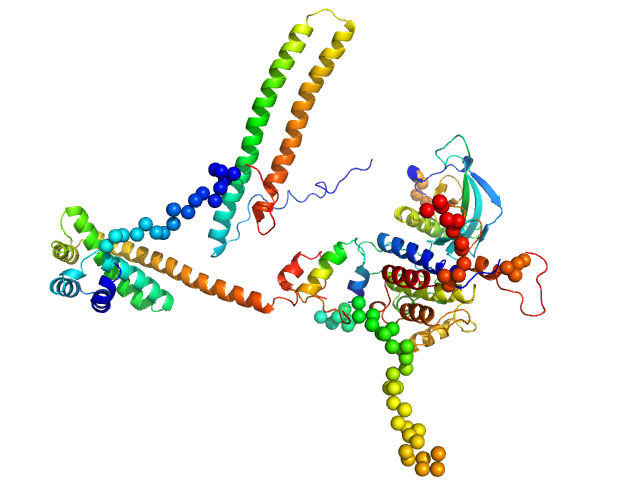
|
| Sample: |
Hsp90 co-chaperone Cdc37 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Disease Variants of FGFR3 Reveal Molecular Basis for the Recognition and Additional Roles for Cdc37 in Hsp90 Chaperone System.
Structure 26(3):446-458.e8 (2018)
Bunney TD, Inglis AJ, Sanfelice D, Farrell B, Kerr CJ, Thompson GS, Masson GR, Thiyagarajan N, Svergun DI, Williams RL, Breeze AL, Katan M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
UniProt ID: Q16543 (None-None) Hsp90 co-chaperone Cdc37
UniProt ID: P22607 (455-768) Fibroblast growth factor receptor 3
|
|
|
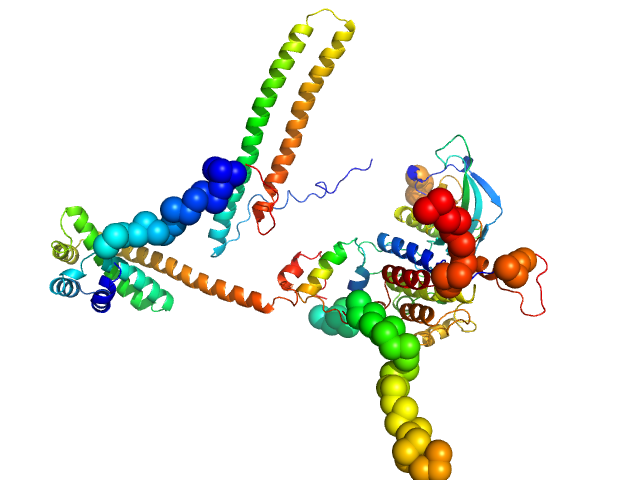
|
| Sample: |
Hsp90 co-chaperone Cdc37 monomer, 44 kDa Homo sapiens protein
Fibroblast growth factor receptor 3 monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Disease Variants of FGFR3 Reveal Molecular Basis for the Recognition and Additional Roles for Cdc37 in Hsp90 Chaperone System.
Structure 26(3):446-458.e8 (2018)
Bunney TD, Inglis AJ, Sanfelice D, Farrell B, Kerr CJ, Thompson GS, Masson GR, Thiyagarajan N, Svergun DI, Williams RL, Breeze AL, Katan M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
UniProt ID: P22607 (455-768) Fibroblast growth factor receptor 3
|
|
|

|
| Sample: |
Fibroblast growth factor receptor 3 monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Disease Variants of FGFR3 Reveal Molecular Basis for the Recognition and Additional Roles for Cdc37 in Hsp90 Chaperone System.
Structure 26(3):446-458.e8 (2018)
Bunney TD, Inglis AJ, Sanfelice D, Farrell B, Kerr CJ, Thompson GS, Masson GR, Thiyagarajan N, Svergun DI, Williams RL, Breeze AL, Katan M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
59 |
nm3 |
|
|