UniProt ID: Q9SZU7 (1-270) Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y)
|
|
|
|
| Sample: |
Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
3.0 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: Q9SZU7 (1-270) Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y)
|
|
|
|
| Sample: |
Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
2.9 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
UniProt ID: Q9SZU7 (1-270) Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y)
|
|
|
|
| Sample: |
Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
5.6 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
494 |
nm3 |
|
|
UniProt ID: Q9SZU7 (1-270) Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y)
|
|
|
|
| Sample: |
Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
5.1 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
403 |
nm3 |
|
|
UniProt ID: Q8IZF6 (26-240) Adhesion G-protein coupled receptor G4
|
|
|
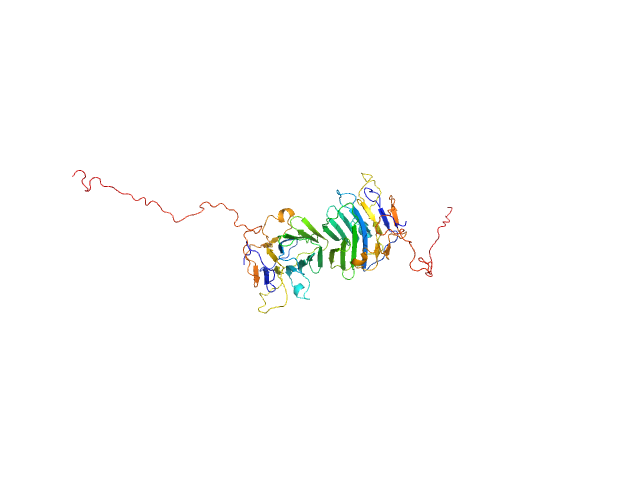
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
2.7 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
UniProt ID: Q8IZF6 (26-240) Adhesion G-protein coupled receptor G4
|
|
|
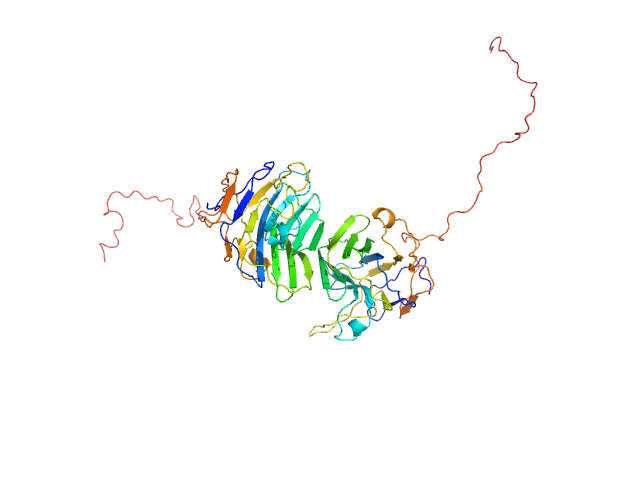
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
UniProt ID: Q8IZF6 (26-240) Adhesion G-protein coupled receptor G4
|
|
|
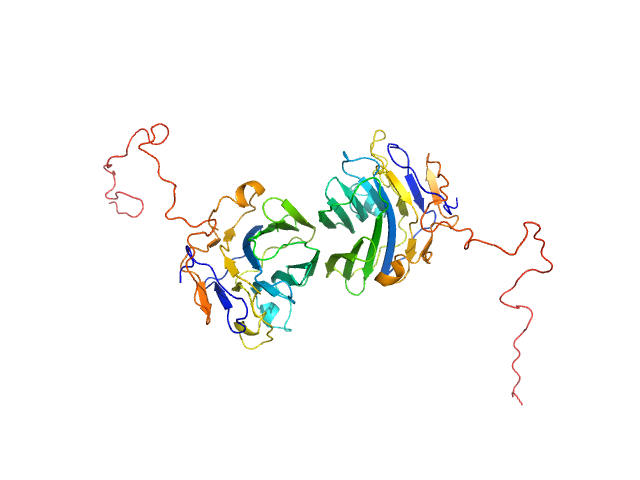
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: Q8IZF6 (26-240) Adhesion G-protein coupled receptor G4
|
|
|
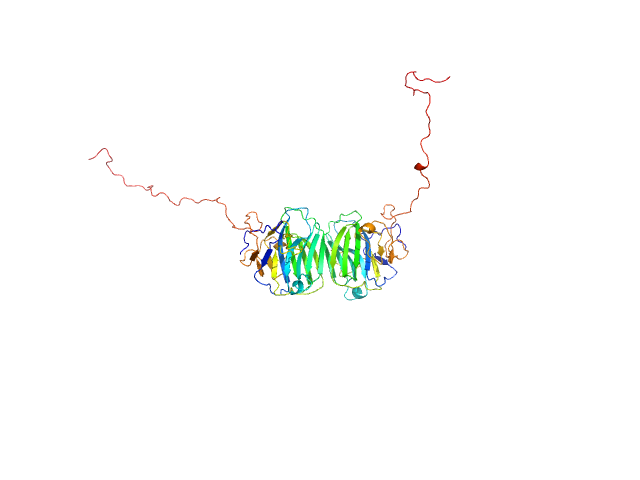
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
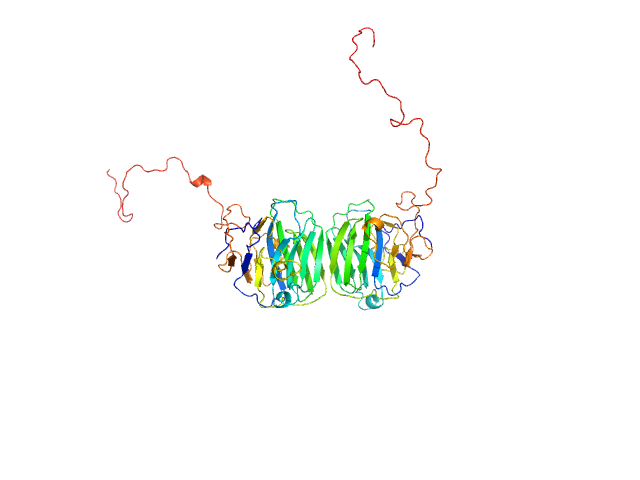
UniProt ID: Q8IZF6 (26-240) Adhesion G-protein coupled receptor G4
|
|
|
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
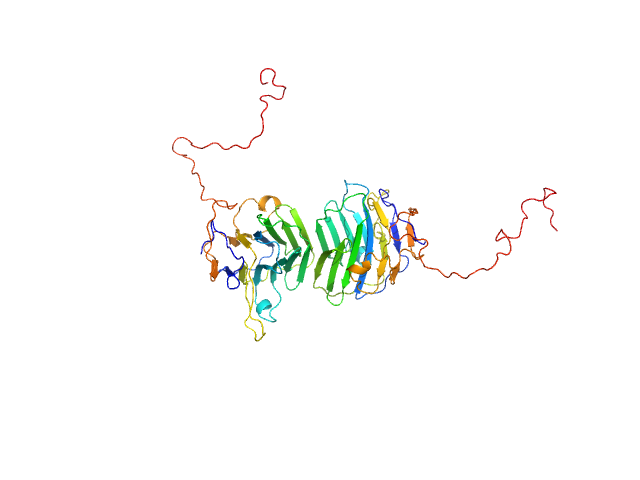
UniProt ID: Q8IZF6 (26-240) Adhesion G-protein coupled receptor G4
|
|
|
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
77 |
nm3 |
|
|