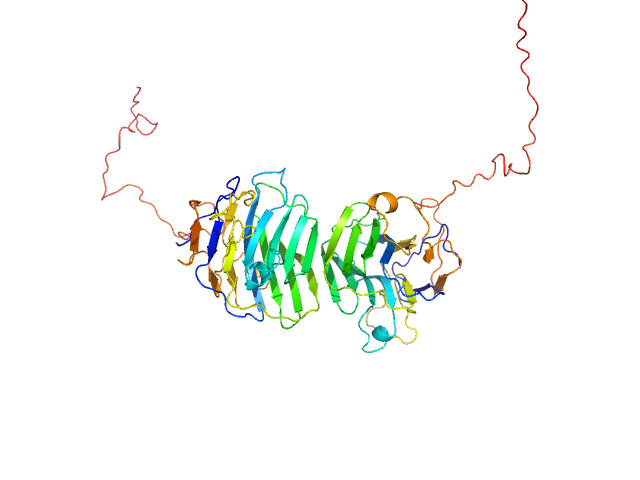
UniProt ID: Q8IZF6 (26-240) Adhesion G-protein coupled receptor G4
|
|
|
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
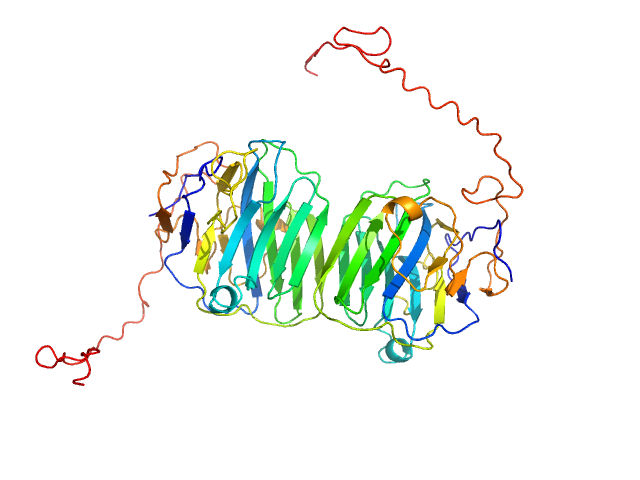
UniProt ID: Q8IZF6 (26-240) Adhesion G-protein coupled receptor G4
|
|
|
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
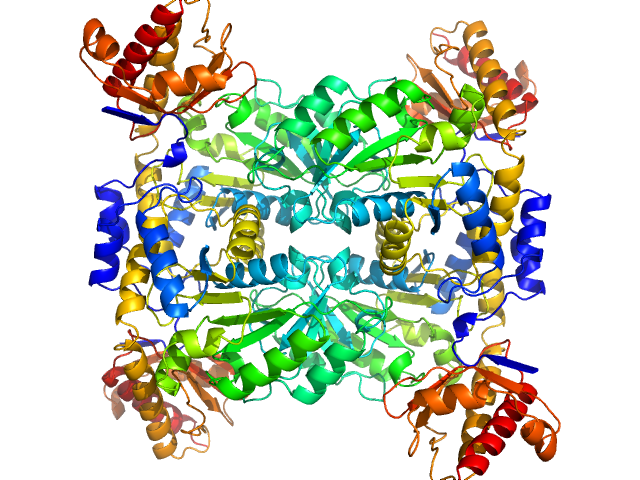
UniProt ID: Q6XPS7 (41-400) L-threonine aldolase
|
|
|
|
| Sample: |
L-threonine aldolase tetramer, 165 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Feb 14
|
One substrate many enzymes virtual screening uncovers missing genes of carnitine biosynthesis in human and mouse.
Nat Commun 15(1):3199 (2024)
Malatesta M, Fornasier E, Di Salvo ML, Tramonti A, Zangelmi E, Peracchi A, Secchi A, Polverini E, Giachin G, Battistutta R, Contestabile R, Percudani R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
209 |
nm3 |
|
|
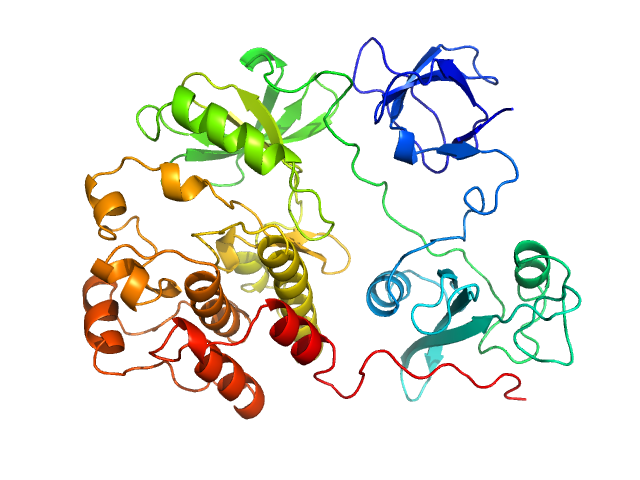
UniProt ID: P12931 (84-536) Proto-oncogene tyrosine-protein kinase Src
|
|
|
|
| Sample: |
Proto-oncogene tyrosine-protein kinase Src monomer, 52 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 1% glycerol and 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 8
|
An allosteric switch between the activation loop and a c-terminal palindromic phospho-motif controls c-Src function
Nature Communications 14(1) (2023)
Cuesta-Hernández H, Contreras J, Soriano-Maldonado P, Sánchez-Wandelmer J, Yeung W, Martín-Hurtado A, Muñoz I, Kannan N, Llimargas M, Muñoz J, Plaza-Menacho I
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
UniProt ID: P20509 (2-1006) DNA polymerase
UniProt ID: P20995 (1-426) DNA polymerase processivity factor component OPG148
UniProt ID: P20536 (1-218) Uracil-DNA glycosylase
|
|
|
|
| Sample: |
DNA polymerase monomer, 117 kDa Vaccinia virus (strain … protein
DNA polymerase processivity factor component OPG148 monomer, 49 kDa Vaccinia virus (strain … protein
Uracil-DNA glycosylase monomer, 27 kDa Vaccinia virus (strain … protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl and 1 mM TCEP, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 25
|
Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus.
PLoS Pathog 20(5):e1011652 (2024)
Burmeister WP, Boutin L, Balestra AC, Gröger H, Ballandras-Colas A, Hutin S, Kraft C, Grimm C, Böttcher B, Fischer U, Tarbouriech N, Iseni F
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
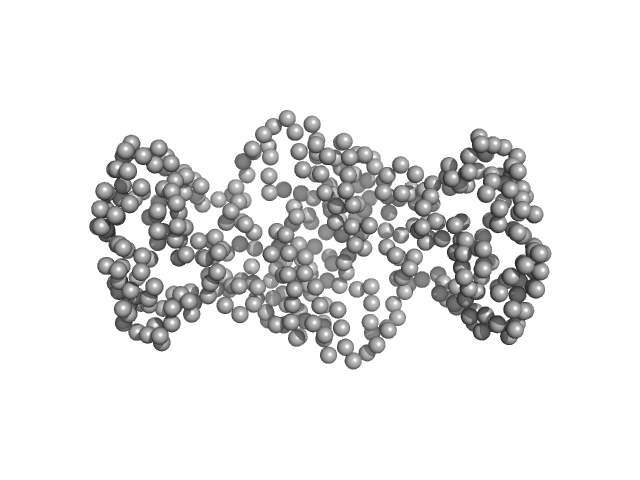
UniProt ID: Q7AL75 (2-182) 20 kDa accessory protein
|
|
|
|
| Sample: |
20 kDa accessory protein dimer, 42 kDa Bacillus thuringiensis serovar … protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2023 Aug 15
|
20-kDa accessory protein (P20) from Bacillus thuringiensis subsp. israelensis ISPC-12: Purification, characterization, solution scattering and structural analysis
International Journal of Biological Macromolecules :127985 (2023)
Kinkar O, Singh R, Prashar A, Kumar A, Hire R, Makde R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
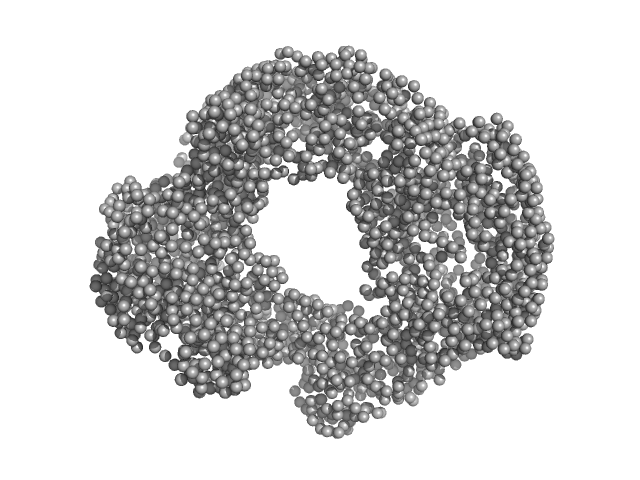
UniProt ID: A0A150M835 (17-670) Acylamino-acid-releasing enzyme (I277L, V491A)
|
|
|
|
| Sample: |
Acylamino-acid-releasing enzyme (I277L, V491A) tetramer, 308 kDa Geobacillus stearothermophilus protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2023 Aug 11
|
Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
FEBS Lett (2024)
Chandravanshi K, Singh R, Bhange GN, Kumar A, Yadav P, Kumar A, Makde RD
|
| RgGuinier |
5.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
424 |
nm3 |
|
|
UniProt ID: P20823 (83-279) Hepatocyte nuclear factor 1-alpha
|
|
|
|
| Sample: |
Hepatocyte nuclear factor 1-alpha monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 1 mM TCEP,, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 25
|
Structural properties of the HNF-1A transactivation domain
Frontiers in Molecular Biosciences 10 (2023)
Kind L, Driver M, Raasakka A, Onck P, Njølstad P, Arnesen T, Kursula P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
UniProt ID: P20823 (83-631) Hepatocyte nuclear factor 1-alpha
|
|
|
|
| Sample: |
Hepatocyte nuclear factor 1-alpha monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
4 mM Tris, 100 mM NaCl, 1 mM TCEP,, pH: 8.5 |
| Experiment: |
SAXS
data collected at CoSAXS, MAX IV on 2022 Apr 2
|
Structural properties of the HNF-1A transactivation domain
Frontiers in Molecular Biosciences 10 (2023)
Kind L, Driver M, Raasakka A, Onck P, Njølstad P, Arnesen T, Kursula P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
109 |
nm3 |
|
|
UniProt ID: P0DTD1 (4254-4392) Replicase polyprotein 1ab (Non-structural protein 10)
|
|
|
|
| Sample: |
Replicase polyprotein 1ab (Non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
|
| Buffer: |
10 mM HEPES, 300 mM NaCl, 1.5% (v/v) glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Nov 25
|
Oligomeric State of β-Coronavirus Non-Structural Protein 10 Stimulators Studied by Small Angle X-ray Scattering
International Journal of Molecular Sciences 24(17):13649 (2023)
Knecht W, Fisher S, Lou J, Sele C, Ma S, Rasmussen A, Pinotsis N, Kozielski F
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
21 |
nm3 |
|
|