UniProt ID: P01558 (1-319) Heat-labile enterotoxin B chain
|
|
|

|
| Sample: |
Heat-labile enterotoxin B chain monomer, 36 kDa Clostridium perfringens protein
|
| Buffer: |
10 mM HEPES, 100 mM NaCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 18
|
Structural Basis of Clostridium perfringens Enterotoxin Activation and Oligomerization by Trypsin
Toxins 15(11):637 (2023)
Ogbu C, Kapoor S, Vecchio A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: P01558 (26-319) Heat-labile enterotoxin B chain
|
|
|

|
| Sample: |
Heat-labile enterotoxin B chain monomer, 33 kDa Clostridium perfringens protein
|
| Buffer: |
10 mM HEPES, 100 mM NaCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 18
|
Structural Basis of Clostridium perfringens Enterotoxin Activation and Oligomerization by Trypsin
Toxins 15(11):637 (2023)
Ogbu C, Kapoor S, Vecchio A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
1 mol% ethylammonium nitrate, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 27
|
Scattering approaches to unravel protein solution behaviors in ionic liquids and deep eutectic solvents: From basic principles to recent developments
Advances in Colloid and Interface Science :103242 (2024)
Han Q, Veríssimo N, Bryant S, Martin A, Huang Y, Pereira J, Santos-Ebinuma V, Zhai J, Bryant G, Drummond C, Greaves T
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
1 mol% ethylammonium nitrate, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 27
|
Scattering approaches to unravel protein solution behaviors in ionic liquids and deep eutectic solvents: From basic principles to recent developments
Advances in Colloid and Interface Science :103242 (2024)
Han Q, Veríssimo N, Bryant S, Martin A, Huang Y, Pereira J, Santos-Ebinuma V, Zhai J, Bryant G, Drummond C, Greaves T
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
1 mol% ethylammonium nitrate, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 27
|
Scattering approaches to unravel protein solution behaviors in ionic liquids and deep eutectic solvents: From basic principles to recent developments
Advances in Colloid and Interface Science :103242 (2024)
Han Q, Veríssimo N, Bryant S, Martin A, Huang Y, Pereira J, Santos-Ebinuma V, Zhai J, Bryant G, Drummond C, Greaves T
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
UniProt ID: None (None-None) Oplophorus-luciferin 2-monooxygenase catalytic subunit
|
|
|
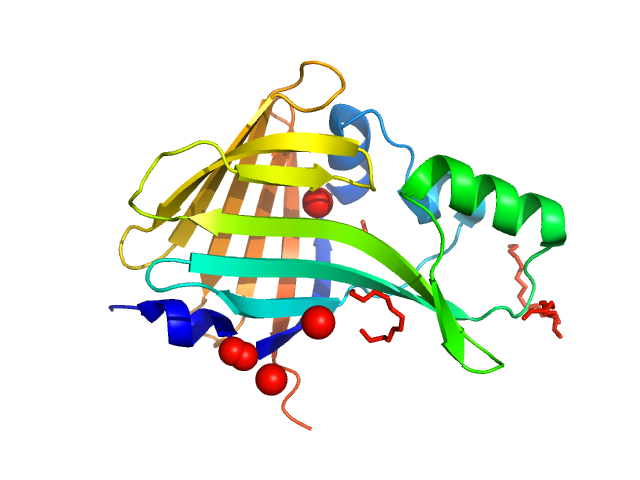
|
| Sample: |
Oplophorus-luciferin 2-monooxygenase catalytic subunit monomer, 20 kDa Oplophorus gracilirostris protein
|
| Buffer: |
10 mM Tris-HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2021 May 27
|
Illuminating the mechanism and allosteric behavior of NanoLuc luciferase.
Nat Commun 14(1):7864 (2023)
Nemergut M, Pluskal D, Horackova J, Sustrova T, Tulis J, Barta T, Baatallah R, Gagnot G, Novakova V, Majerova M, Sedlackova K, Marques SM, Toul M, Damborsky J, Prokop Z, Bednar D, Janin YL, Marek M
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
UniProt ID: I3R177 (1-152) AsnC family transcriptional regulator
|
|
|
|
| Sample: |
AsnC family transcriptional regulator dimer, 36 kDa Haloferax mediterranei (strain … protein
|
| Buffer: |
50 mM Tris, 1.5 M NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 12
|
Global Lrp regulator protein from Haloferax mediterranei: Transcriptional analysis and structural characterization.
Int J Biol Macromol :129541 (2024)
Matarredona L, García-Bonete MJ, Guío J, Camacho M, Fillat MF, Esclapez J, Bonete MJ
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: Q43349 (176-254) 29 kDa ribonucleoprotein, chloroplastic
|
|
|
|
| Sample: |
29 kDa ribonucleoprotein, chloroplastic monomer, 8 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM sodium phosphate, 30 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2021 Oct 19
|
A prion-like domain is required for phase separation and chloroplast RNA processing during cold acclimation in Arabidopsis.
Plant Cell (2024)
Legen J, Lenzen B, Kachariya N, Feltgen S, Gao Y, Mergenthal S, Weber W, Klotzsch E, Zoschke R, Sattler M, Schmitz-Linneweber C
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
UniProt ID: Q43349 (176-254) 29 kDa ribonucleoprotein, chloroplastic
|
|
|
|
| Sample: |
29 kDa ribonucleoprotein, chloroplastic monomer, 8 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM sodium phosphate, 30 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2021 Oct 19
|
A prion-like domain is required for phase separation and chloroplast RNA processing during cold acclimation in Arabidopsis.
Plant Cell (2024)
Legen J, Lenzen B, Kachariya N, Feltgen S, Gao Y, Mergenthal S, Weber W, Klotzsch E, Zoschke R, Sattler M, Schmitz-Linneweber C
|
| RgGuinier |
2.4 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
UniProt ID: P02925 (25-296) Ribose import binding protein RbsB
|
|
|
|
| Sample: |
Ribose import binding protein RbsB monomer, 30 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 10% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 9
|
SAXS/WAXS data of conformationally flexible ribose binding protein
Data in Brief :109932 (2023)
Choudhury J, Yonezawa K, Anu A, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
36 |
nm3 |
|
|