UniProt ID: P02925 (25-296) Ribose import binding protein RbsB
UniProt ID: None (None-None) Ribose
|
|
|
|
| Sample: |
Ribose import binding protein RbsB monomer, 30 kDa Escherichia coli (strain … protein
Ribose monomer, 0 kDa
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 10% glycerol, 1 mM ribose, pH: 7 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 9
|
SAXS/WAXS data of conformationally flexible ribose binding protein
Data in Brief :109932 (2023)
Choudhury J, Yonezawa K, Anu A, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
UniProt ID: P02925 (25-296) Ribose import binding protein RbsB
UniProt ID: None (None-None) Ribose
|
|
|
|
| Sample: |
Ribose import binding protein RbsB monomer, 30 kDa Escherichia coli (strain … protein
Ribose monomer, 0 kDa
|
| Buffer: |
Tris 50 mM, 50 mM NaCl, 10% glycerol, 5 mM ribose, pH: 7 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 9
|
SAXS/WAXS data of conformationally flexible ribose binding protein
Data in Brief :109932 (2023)
Choudhury J, Yonezawa K, Anu A, Shimizu N, Chaudhuri B
|
| RgGuinier |
1.9 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
UniProt ID: Q9NYQ6 (557-995) Cadherin EGF LAG seven-pass G-type receptor 1
|
|
|
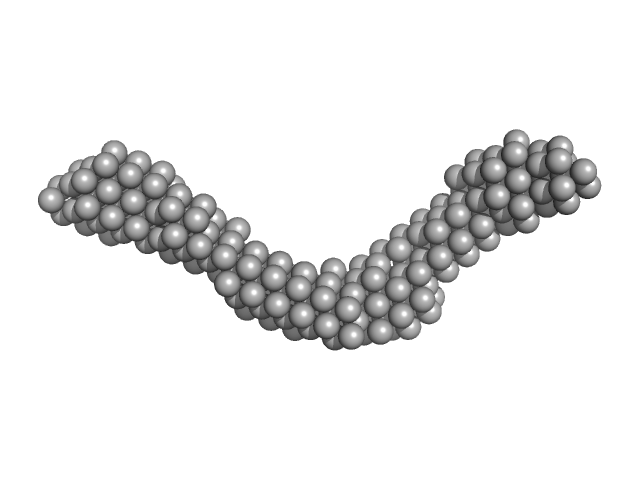
|
| Sample: |
Cadherin EGF LAG seven-pass G-type receptor 1 monomer, 49 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM KCl, 50 mM NaCl, 2 mM CaCl2, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Jul 1
|
CELSR1, a core planar cell polarity protein, features a weakly adhesive and flexible cadherin ectodomain.
Structure (2024)
Tamilselvan E, Sotomayor M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
UniProt ID: A0A0A7LTX5 (48-342) Biofilm regulatory protein
|
|
|
|
| Sample: |
Biofilm regulatory protein monomer, 34 kDa Streptococcus dysgalactiae subsp. … protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 5 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 1
|
Structural insights of an LCP protein-LytR-from Streptococcus dysgalactiae subs. dysgalactiae through biophysical and in silico methods.
Front Chem 12:1379914 (2024)
Paquete-Ferreira J, Freire F, Fernandes HS, Muthukumaran J, Ramos J, Bryton J, Panjkovich A, Svergun D, Santos MFA, Correia MAS, Fernandes AR, Romão MJ, Sousa SF, Santos-Silva T
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
UniProt ID: A0A0A7LTX5 (48-342) Biofilm regulatory protein
|
|
|
|
| Sample: |
Biofilm regulatory protein monomer, 34 kDa Streptococcus dysgalactiae subsp. … protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 5 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 1
|
Structural insights of an LCP protein-LytR-from Streptococcus dysgalactiae subs. dysgalactiae through biophysical and in silico methods.
Front Chem 12:1379914 (2024)
Paquete-Ferreira J, Freire F, Fernandes HS, Muthukumaran J, Ramos J, Bryton J, Panjkovich A, Svergun D, Santos MFA, Correia MAS, Fernandes AR, Romão MJ, Sousa SF, Santos-Silva T
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
UniProt ID: P09936 (1-223) Ubiquitin carboxyl-terminal hydrolase isozyme L1
|
|
|
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase isozyme L1 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.03% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 Sep 2
|
Altered protein dynamics and a more reactive catalytic cysteine in a neurodegeneration-associated UCHL1 mutant.
J Mol Biol :168438 (2024)
Kenny S, Lai CH, Chiang TS, Brown K, Hewitt CS, Krabill AD, Chang HT, Wang YS, Flaherty DP, Danny Hsu ST, Das C
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
UniProt ID: P09936 (1-223) Ubiquitin carboxyl-terminal hydrolase isozyme L1 (R178Q)
|
|
|
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase isozyme L1 (R178Q) monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.03% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 Sep 2
|
Altered protein dynamics and a more reactive catalytic cysteine in a neurodegeneration-associated UCHL1 mutant.
J Mol Biol :168438 (2024)
Kenny S, Lai CH, Chiang TS, Brown K, Hewitt CS, Krabill AD, Chang HT, Wang YS, Flaherty DP, Danny Hsu ST, Das C
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
UniProt ID: Q9NRC8-1 (1-400) Isoform 1 of NAD-dependent protein deacetylase sirtuin-7
|
|
|
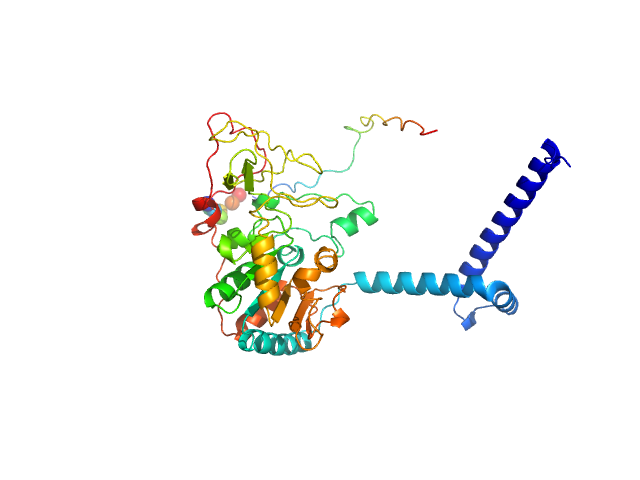
|
| Sample: |
Isoform 1 of NAD-dependent protein deacetylase sirtuin-7 monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 16
|
Substrates and Cyclic Peptide Inhibitors of the Oligonucleotide-Activated Sirtuin 7.
Angew Chem Int Ed Engl :e202314597 (2023)
Bolding JE, Nielsen AL, Jensen I, Hansen TN, Ryberg LA, Jameson ST, Harris P, Peters GHJ, Denu JM, Rogers JM, Olsen CA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
UniProt ID: P30153 (1-589) Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform
UniProt ID: Q14738-1 (1-602) Isoform Delta-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform (R160H)
UniProt ID: P67775-1 (1-309) Isoform 1 of Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform
|
|
|
|
| Sample: |
Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform monomer, 65 kDa Homo sapiens protein
Isoform Delta-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform (R160H) monomer, 70 kDa Homo sapiens protein
Isoform 1 of Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 50 µM MnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 20
|
B56δ long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc Natl Acad Sci U S A 121(1):e2310727120 (2024)
Wu CG, Balakrishnan VK, Merrill RA, Parihar PS, Konovolov K, Chen YC, Xu Z, Wei H, Sundaresan R, Cui Q, Wadzinski BE, Swingle MR, Musiyenko A, Chung WK, Honkanen RE, Suzuki A, Huang X, Strack S, Xing Y
|
| RgGuinier |
4.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
266 |
nm3 |
|
|
UniProt ID: P30153 (1-589) Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform
UniProt ID: P67775-1 (1-309) Isoform 1 of Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform
UniProt ID: Q14738-1 (1-602) Isoform Delta-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform (R160H, E198K)
|
|
|
|
| Sample: |
Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform monomer, 65 kDa Homo sapiens protein
Isoform 1 of Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform monomer, 36 kDa Homo sapiens protein
Isoform Delta-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform (R160H, E198K) monomer, 70 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 50 µM MnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 20
|
B56δ long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc Natl Acad Sci U S A 121(1):e2310727120 (2024)
Wu CG, Balakrishnan VK, Merrill RA, Parihar PS, Konovolov K, Chen YC, Xu Z, Wei H, Sundaresan R, Cui Q, Wadzinski BE, Swingle MR, Musiyenko A, Chung WK, Honkanen RE, Suzuki A, Huang X, Strack S, Xing Y
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
305 |
nm3 |
|
|