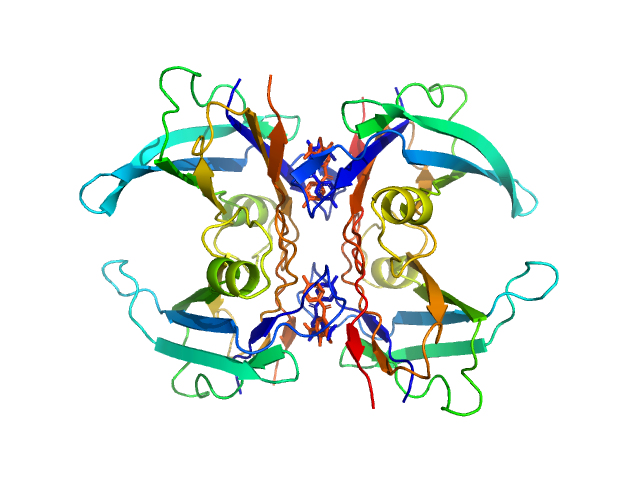
UniProt ID: P02766 (21-147) Transthyretin
UniProt ID: None (None-None) Tolcapone
|
|
|
|
| Sample: |
Transthyretin tetramer, 55 kDa Homo sapiens protein
Tolcapone dimer, 1 kDa
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 19
|
Differentiating the solution structures and stability of transthyretin tetramer complexed with tolcapone and tafamidis using SEC-SWAXS and NMR.
J Appl Crystallogr 58(Pt 4):1373-1383 (2025)
Shih O, Feng YC, Agrawal S, Liao KF, Yeh YQ, Chang JW, Yu TY, Jeng US
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
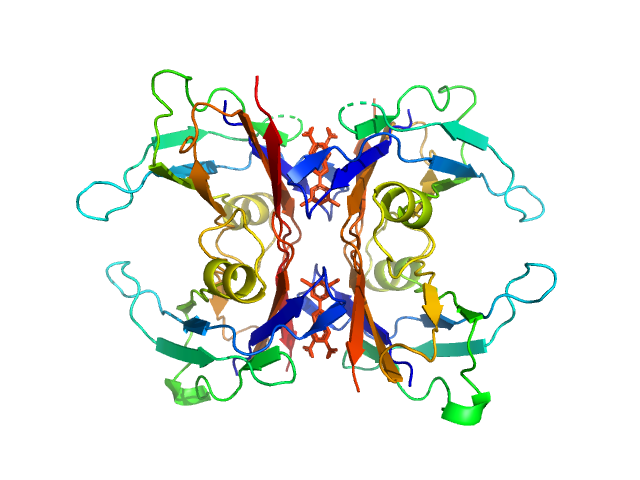
UniProt ID: P02766 (21-147) Transthyretin
UniProt ID: None (None-None) Tafamidis
|
|
|
|
| Sample: |
Transthyretin tetramer, 55 kDa Homo sapiens protein
Tafamidis dimer, 1 kDa
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 19
|
Differentiating the solution structures and stability of transthyretin tetramer complexed with tolcapone and tafamidis using SEC-SWAXS and NMR.
J Appl Crystallogr 58(Pt 4):1373-1383 (2025)
Shih O, Feng YC, Agrawal S, Liao KF, Yeh YQ, Chang JW, Yu TY, Jeng US
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
UniProt ID: P33292 (1-576) Peroxisomal targeting signal receptor
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDX84_fit1_model1.png)
|
| Sample: |
Peroxisomal targeting signal receptor monomer, 65 kDa Komagataella pastoris protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 3% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 May 15
|
Structure of Pex8 in complex with peroxisomal receptor Pex5 reveals its essential role in peroxisomal cargo translocation
(2025)
Ekal L, Wendscheck D, David Y, Chojnowski G, Jeffries C, Mullapudi E, Schuldiner M, Warscheid B, Zalckvar E, Wilmanns M
|
| RgGuinier |
5.0 |
nm |
| Dmax |
21.4 |
nm |
| VolumePorod |
131 |
nm3 |
|
|
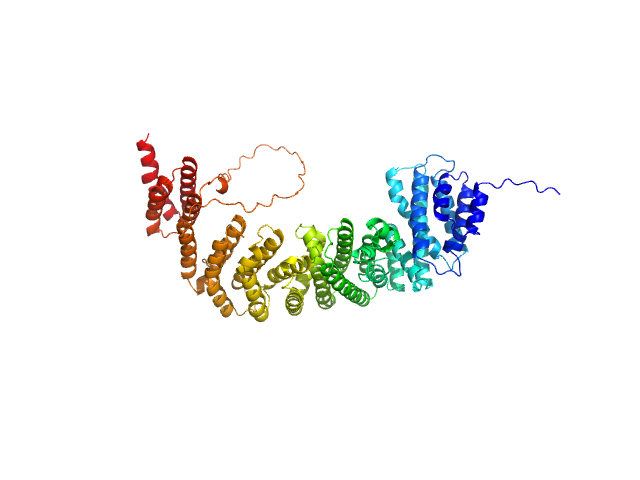
UniProt ID: Q01962 (33-713) Peroxisomal biogenesis factor 8
|
|
|
|
| Sample: |
Peroxisomal biogenesis factor 8 monomer, 78 kDa Komagataella pastoris protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 3% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 17
|
Structure of Pex8 in complex with peroxisomal receptor Pex5 reveals its essential role in peroxisomal cargo translocation
(2025)
Ekal L, Wendscheck D, David Y, Chojnowski G, Jeffries C, Mullapudi E, Schuldiner M, Warscheid B, Zalckvar E, Wilmanns M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
UniProt ID: P33292 (1-576) Peroxisomal targeting signal receptor
UniProt ID: Q01962 (33-713) Peroxisomal biogenesis factor 8
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDXA4_fit1_model1.png)
|
| Sample: |
Peroxisomal targeting signal receptor monomer, 65 kDa Komagataella pastoris protein
Peroxisomal biogenesis factor 8 monomer, 78 kDa Komagataella pastoris protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 3% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 17
|
Structure of Pex8 in complex with peroxisomal receptor Pex5 reveals its essential role in peroxisomal cargo translocation
(2025)
Ekal L, Wendscheck D, David Y, Chojnowski G, Jeffries C, Mullapudi E, Schuldiner M, Warscheid B, Zalckvar E, Wilmanns M
|
| RgGuinier |
5.1 |
nm |
| Dmax |
23.5 |
nm |
| VolumePorod |
247 |
nm3 |
|
|
UniProt ID: Q01962 (33-713) Peroxisomal biogenesis factor 8
UniProt ID: P33292 (1-276) Peroxisomal targeting signal receptor
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDXB4_fit1_model1.png)
|
| Sample: |
Peroxisomal biogenesis factor 8 monomer, 78 kDa Komagataella pastoris protein
Peroxisomal targeting signal receptor monomer, 32 kDa Komagataella pastoris protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 3% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 17
|
Structure of Pex8 in complex with peroxisomal receptor Pex5 reveals its essential role in peroxisomal cargo translocation
(2025)
Ekal L, Wendscheck D, David Y, Chojnowski G, Jeffries C, Mullapudi E, Schuldiner M, Warscheid B, Zalckvar E, Wilmanns M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
177 |
nm3 |
|
|
UniProt ID: Q01962 (33-713) Peroxisomal biogenesis factor 8
UniProt ID: P33292 (259-576) Peroxisomal targeting signal receptor
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDXC4_fit1_model1.png)
|
| Sample: |
Peroxisomal biogenesis factor 8 monomer, 78 kDa Komagataella pastoris protein
Peroxisomal targeting signal receptor monomer, 36 kDa Komagataella pastoris protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 3% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 17
|
Structure of Pex8 in complex with peroxisomal receptor Pex5 reveals its essential role in peroxisomal cargo translocation
(2025)
Ekal L, Wendscheck D, David Y, Chojnowski G, Jeffries C, Mullapudi E, Schuldiner M, Warscheid B, Zalckvar E, Wilmanns M
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
141 |
nm3 |
|
|
UniProt ID: P23246 (214-707) Splicing factor, proline- and glutamine-rich
|
|
|
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
|
| Buffer: |
150 mM KCl, 1.5% glycerol, 20 mM HEPES, 1 mM DTT (in 100% H2O), pH: 7.4 |
| Experiment: |
SANS
data collected at Quokka - Small Angle Neutron Scattering, Australian Centre for Neutron Scattering (ANSTO) on 2023 Jan 23
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
8.7 |
nm |
| Dmax |
30.7 |
nm |
| VolumePorod |
241000 |
nm3 |
|
|
UniProt ID: Q9AES2 (31-1167) IgA protease
|
|
|
|
| Sample: |
IgA protease monomer, 127 kDa Thomasclavelia ramosa protein
|
| Buffer: |
50 mM Tris-HCl pH 7.5, 100 mM NaCl, 2% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Apr 19
|
Molecular basis of Fab-dependent IgA antibody recognition by gut-bacterial metallopeptidases
The EMBO Journal (2025)
Márquez-Moñino M, Martínez Gascueña A, Azzam T, Persson A, Manzanares-Gomez A, Aguillo-Urarte M, Brown T, Montero-Sagarminaga A, Lood R, Naegeli A, Connell S, Sastre D, Sundberg E, Trastoy B
|
| RgGuinier |
6.4 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
208 |
nm3 |
|
|
UniProt ID: Q9AES2 (31-878) IgA protease
|
|
|
|
| Sample: |
IgA protease monomer, 96 kDa Thomasclavelia ramosa protein
|
| Buffer: |
50 mM Tris-HCl pH 7.5, 100 mM NaCl, 2% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Apr 19
|
Molecular basis of Fab-dependent IgA antibody recognition by gut-bacterial metallopeptidases
The EMBO Journal (2025)
Márquez-Moñino M, Martínez Gascueña A, Azzam T, Persson A, Manzanares-Gomez A, Aguillo-Urarte M, Brown T, Montero-Sagarminaga A, Lood R, Naegeli A, Connell S, Sastre D, Sundberg E, Trastoy B
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
101 |
nm3 |
|
|