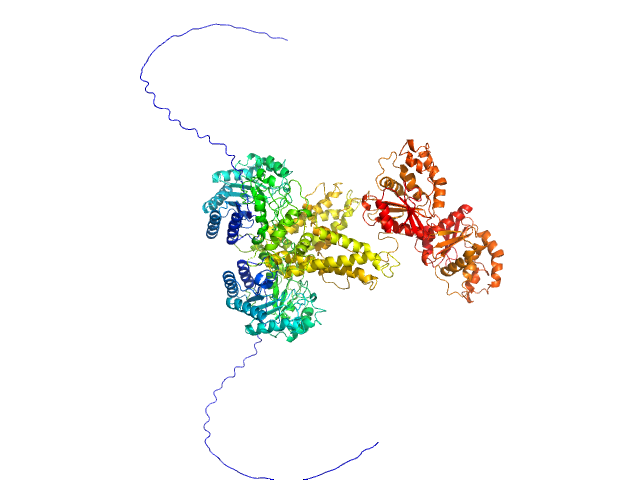
UniProt ID: O60502 (11-766) Protein O-GlcNAcase
|
|
|
|
| Sample: |
Protein O-GlcNAcase dimer, 163 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris buffer, 150 mM NaCl, and 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2023 Jun 22
|
Multi-domain O-GlcNAcase structures reveal allosteric regulatory mechanisms
Jan Skov Pedersen
|
| RgGuinier |
5.4 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
443 |
nm3 |
|
|
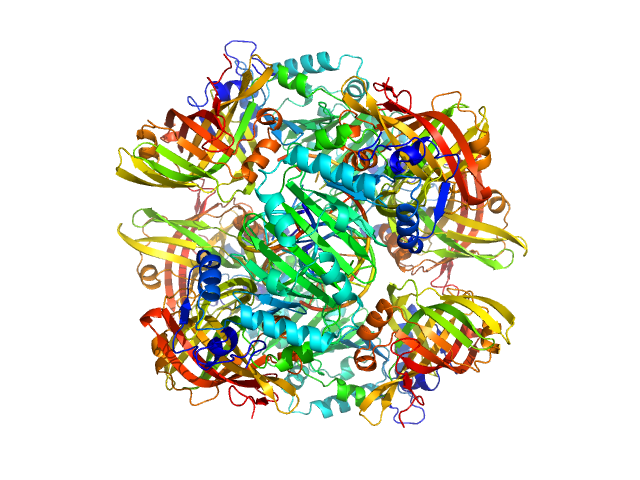
UniProt ID: P0DTD1 (6465-6798) Non structural Protein 15
|
|
|
|
| Sample: |
Non structural Protein 15 monomer, 251 kDa Severe acute respiratory … protein
|
| Buffer: |
20mM HEPES, 150mM Nacl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2024 Jun 14
|
Non Structural Protein 15 in Complex with RNA 9mer Oligo of SARS CoV 2
Hira Singh Gariya
|
| RgGuinier |
4.7 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
356 |
nm3 |
|
|
UniProt ID: A0A0H2URK1 (187-385) Functional binding region (187-385) of the pneumococcal serine-rich repeat protein
|
|
|
|
| Sample: |
Functional binding region (187-385) of the pneumococcal serine-rich repeat protein monomer, 22 kDa Streptococcus pneumoniae protein
|
| Buffer: |
PBS 5 % Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Feb 27
|
The BR domain of PsrP interacts with extracellular DNA to promote bacterial aggregation; structural insights into pneumococcal biofilm formation.
Sci Rep 6:32371 (2016)
Schulte T, Mikaelsson C, Beaussart A, Kikhney A, Deshmukh M, Wolniak S, Pathak A, Ebel C, Löfling J, Fogolari F, Henriques-Normark B, Dufrêne YF, Svergun D, Nygren PÅ, Achour A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
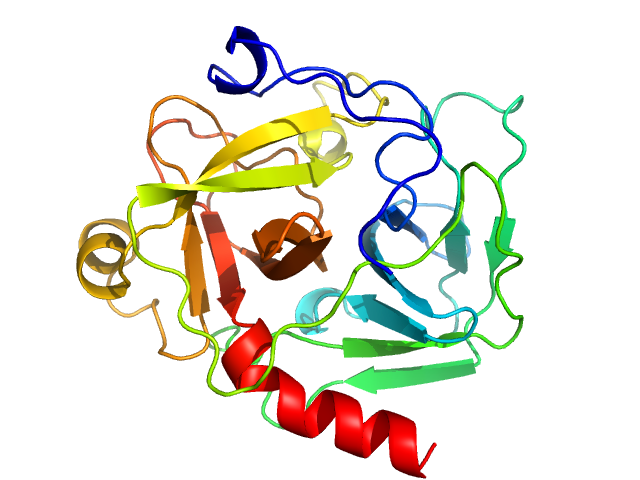
UniProt ID: P00766 (None-None) Chymotrypsinogen A
|
|
|
|
| Sample: |
Chymotrypsinogen A monomer, 26 kDa Bos taurus protein
|
| Buffer: |
100 mM Tris/HCl 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 19
|
Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering
Journal of Applied Crystallography 40(s1):s245-s249 (2007)
Mylonas E, Svergun D
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.0 |
nm |
|
|
UniProt ID: M4N8T6 (None-None) S-layer protein
|
|
|

|
| Sample: |
S-layer protein monomer, 116 kDa Lysinibacillus sphaericus protein
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Analysis of self-assembly of S-layer protein slp-B53 from Lysinibacillus sphaericus.
Eur Biophys J 46(1):77-89 (2017)
Liu J, Falke S, Drobot B, Oberthuer D, Kikhney A, Guenther T, Fahmy K, Svergun D, Betzel C, Raff J
|
| RgGuinier |
5.8 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
495 |
nm3 |
|
|
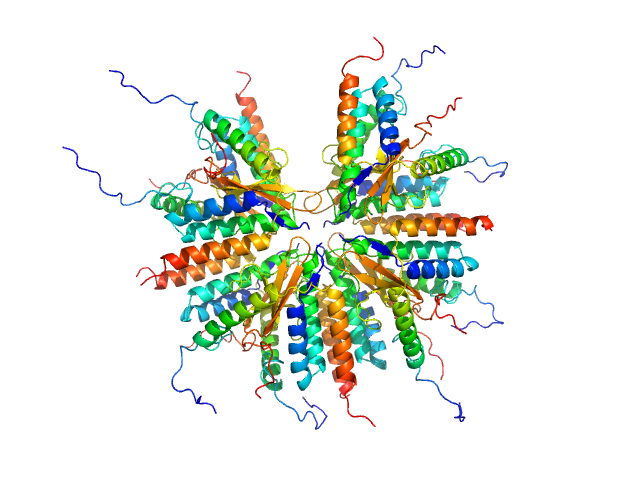
UniProt ID: A0A0D7C2L1 (1-92) Plasmid stabilization protein ParE
UniProt ID: A0A0F6F6Q9 (14-75) Uncharacterized protein (Antitoxin)
|
|
|
|
| Sample: |
Plasmid stabilization protein ParE 16-mer, 188 kDa Escherichia coli protein
Uncharacterized protein (Antitoxin) 16-mer, 135 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCl 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Dec 9
|
A unique hetero-hexadecameric architecture displayed by the Escherichia coli O157 PaaA2-ParE2 antitoxin-toxin complex.
J Mol Biol 428(8):1589-603 (2016)
Sterckx YG, Jové T, Shkumatov AV, Garcia-Pino A, Geerts L, De Kerpel M, Lah J, De Greve H, Van Melderen L, Loris R
|
| RgGuinier |
3.8 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
312 |
nm3 |
|
|
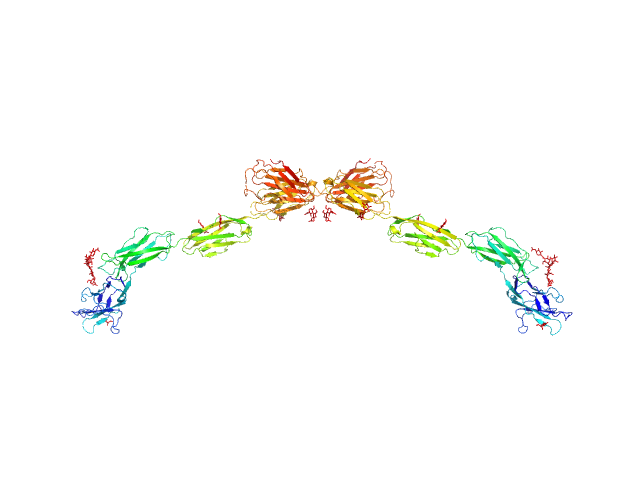
UniProt ID: P20917 (20-508) Myelin-associated glycoprotein Ig domains 1-5
|
|
|
|
| Sample: |
Myelin-associated glycoprotein Ig domains 1-5 dimer, 108 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
6.8 |
nm |
| Dmax |
23.8 |
nm |
| VolumePorod |
177 |
nm3 |
|
|
UniProt ID: Q15185 (1-160) Prostaglandin E synthase 3
|
|
|
|
| Sample: |
Prostaglandin E synthase 3 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM NaCl, 5 mM B-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2012 Jun 22
|
The C-terminal region of the human p23 chaperone modulates its structure and function.
Arch Biochem Biophys 565:57-67 (2015)
Seraphim TV, Gava LM, Mokry DZ, Cagliari TC, Barbosa LR, Ramos CH, Borges JC
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
UniProt ID: P31483-2 (93-274) Nucleolysin TIA-1 isoform p40
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Jul 2
|
TIA-1 RRM23 binding and recognition of target oligonucleotides.
Nucleic Acids Res 45(8):4944-4957 (2017)
Waris S, García-Mauriño SM, Sivakumaran A, Beckham SA, Loughlin FE, Gorospe M, Díaz-Moreno I, Wilce MCJ, Wilce JA
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
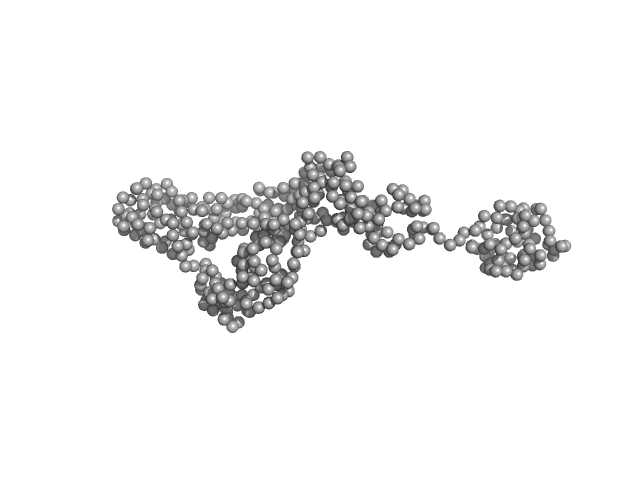
UniProt ID: Q96FI4 (None-None) Endonuclease 8-like 1
|
|
|
|
| Sample: |
Endonuclease 8-like 1 monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
25mM HEPES 300mM NaCl 1mM DTT 10% Glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Mar 13
|
Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase.
Nucleic Acids Res 45(5):2897-2909 (2017)
Prakash A, Moharana K, Wallace SS, Doublié S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
81 |
nm3 |
|
|