UniProt ID: Q9BE39 (None-None) Bovine β Cardiac Myosin S1 fragment
|
|
|
|
| Sample: |
Bovine β Cardiac Myosin S1 fragment monomer, 117 kDa Bos taurus protein
|
| Buffer: |
10 mM HEPES, 50 mM NaCl, 1 mM NaN3, 2.5 mM MgCl2, 2 mM ADP, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Mar 1
|
Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil.
Nat Commun 8(1):190 (2017)
Planelles-Herrero VJ, Hartman JJ, Robert-Paganin J, Malik FI, Houdusse A
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
UniProt ID: Q9BE39 (None-None) Bovine β Cardiac Myosin S1 fragment
|
|
|
|
| Sample: |
Bovine β Cardiac Myosin S1 fragment monomer, 117 kDa Bos taurus protein
|
| Buffer: |
10 mM HEPES, 50 mM NaCl, 1 mM NaN3, 2.5 mM MgCl2, 2 mM ADP, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Mar 1
|
Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil.
Nat Commun 8(1):190 (2017)
Planelles-Herrero VJ, Hartman JJ, Robert-Paganin J, Malik FI, Houdusse A
|
| RgGuinier |
4.0 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
UniProt ID: Q46977 (603-850) RNase E 603-850
|
|
|
|
| Sample: |
RNase E 603-850 monomer, 30 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris HCl, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 10 mM DTT and 5 % glycerol (v/v), pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 5
|
Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes.
Nucleic Acids Res 46(1):387-402 (2018)
Bruce HA, Du D, Matak-Vinkovic D, Bandyra KJ, Broadhurst RW, Martin E, Sobott F, Shkumatov AV, Luisi BF
|
| RgGuinier |
5.3 |
nm |
| Dmax |
27.5 |
nm |
| VolumePorod |
139 |
nm3 |
|
|
UniProt ID: Q46977 (603-850) RNase E 603-850
UniProt ID: W8ZZD4 (1-421) ATP-dependent RNA helicase RhlB
|
|
|

|
| Sample: |
RNase E 603-850 monomer, 30 kDa Escherichia coli protein
ATP-dependent RNA helicase RhlB monomer, 47 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris HCl, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 10 mM DTT and 5 % glycerol (v/v), pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Feb 11
|
Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes.
Nucleic Acids Res 46(1):387-402 (2018)
Bruce HA, Du D, Matak-Vinkovic D, Bandyra KJ, Broadhurst RW, Martin E, Sobott F, Shkumatov AV, Luisi BF
|
| RgGuinier |
5.4 |
nm |
| Dmax |
29.5 |
nm |
| VolumePorod |
183 |
nm3 |
|
|
UniProt ID: Q46977 (603-850) RNase E 603-850
UniProt ID: W8ZZD4 (1-421) ATP-dependent RNA helicase RhlB
UniProt ID: A1AEW7 (1-432) Enolase
|
|
|

|
| Sample: |
RNase E 603-850 monomer, 30 kDa Escherichia coli protein
ATP-dependent RNA helicase RhlB monomer, 47 kDa Escherichia coli protein
Enolase dimer, 91 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris HCl, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 10 mM DTT and 5 % glycerol (v/v), pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Jul 16
|
Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes.
Nucleic Acids Res 46(1):387-402 (2018)
Bruce HA, Du D, Matak-Vinkovic D, Bandyra KJ, Broadhurst RW, Martin E, Sobott F, Shkumatov AV, Luisi BF
|
| RgGuinier |
6.4 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
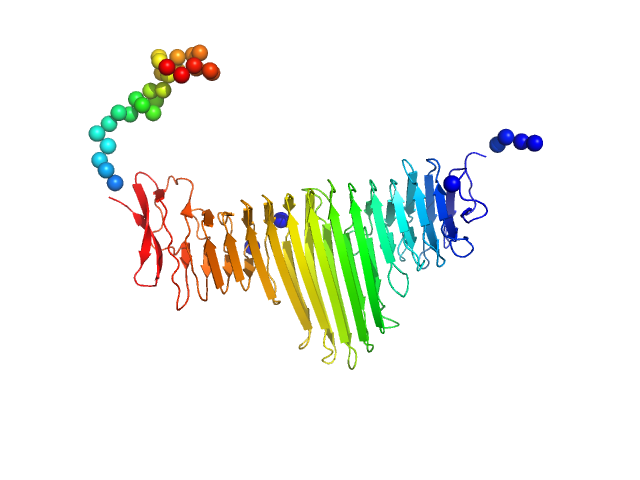
UniProt ID: A0A0H2V5A3 (37-500) Alpha domain of autotransporter protein UpaB
|
|
|
|
| Sample: |
Alpha domain of autotransporter protein UpaB monomer, 48 kDa E. Coli CFT073 protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 May 1
|
Unique structural features of a bacterial autotransporter adhesin suggest mechanisms for interaction with host macromolecules.
Nat Commun 10(1):1967 (2019)
Paxman JJ, Lo AW, Sullivan MJ, Panjikar S, Kuiper M, Whitten AE, Wang G, Luan CH, Moriel DG, Tan L, Peters KM, Phan MD, Gee CL, Ulett GC, Schembri MA, Heras B
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
UniProt ID: Q8I2N9 (None-None) CS domain protein, putative
|
|
|

|
| Sample: |
CS domain protein, putative monomer, 19 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM NaCl, 2 mM EDTA, 1 mM B-mercaptoethanol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2015 May 26
|
Comparative studies of the low-resolution structure of two p23 co-chaperones for Hsp90 identified in Plasmodium falciparum genome.
Int J Biol Macromol 108:193-204 (2018)
Silva NSM, Seraphim TV, Minari K, Barbosa LRS, Borges JC
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.5 |
nm |
|
|
UniProt ID: Q8IKU1 (None-None) Co-chaperone p23
|
|
|

|
| Sample: |
Co-chaperone p23 monomer, 31 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM NaCl, 2 mM EDTA, 1 mM B-mercaptoethanol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2015 May 26
|
Comparative studies of the low-resolution structure of two p23 co-chaperones for Hsp90 identified in Plasmodium falciparum genome.
Int J Biol Macromol 108:193-204 (2018)
Silva NSM, Seraphim TV, Minari K, Barbosa LRS, Borges JC
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
|
|
UniProt ID: A4HIK6 (None-None) SGT protein
|
|
|
|
| Sample: |
SGT protein dimer, 92 kDa Leishmania braziliensis protein
|
| Buffer: |
20 mM Potassium Phosphate, 100 mM KCl, 10 mM EDTA, 1 mM B-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2014 May 14
|
Structural and functional studies of the Leishmania braziliensis SGT co-chaperone indicate that it shares structural features with HIP and can interact with both Hsp90 and Hsp70 with similar affinities.
Int J Biol Macromol 118(Pt A):693-706 (2018)
Coto ALS, Seraphim TV, Batista FAH, Dores-Silva PR, Barranco ABF, Teixeira FR, Gava LM, Borges JC
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.0 |
nm |
|
|
UniProt ID: A4HAD9 (None-None) Leishmania braziliensis p23 isoform B
|
|
|

|
| Sample: |
Leishmania braziliensis p23 isoform B monomer, 23 kDa Leishmania braziliensis protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM NaCl, 5 mM B-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2012 Mar 26
|
Identification of two p23 co-chaperone isoforms in Leishmania braziliensis exhibiting similar structures and Hsp90 interaction properties despite divergent stabilities.
FEBS J 282(2):388-406 (2015)
Batista FA, Almeida GS, Seraphim TV, Silva KP, Murta SM, Barbosa LR, Borges JC
|
| RgGuinier |
3.0 |
nm |
| Dmax |
13.0 |
nm |
|
|