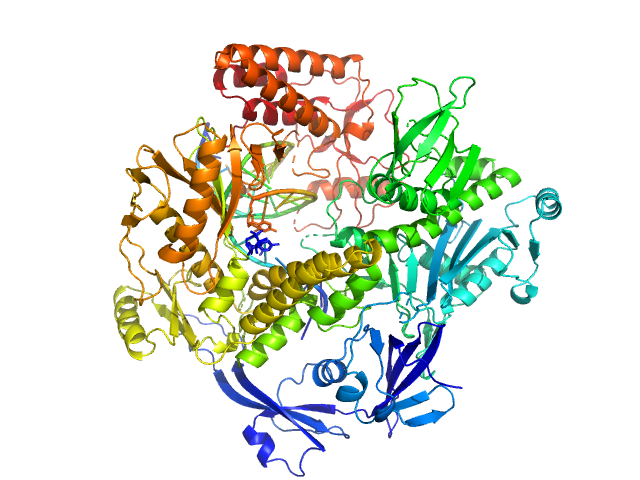
UniProt ID: P20509 (1-1006) DNA polymerase E9 exo- mutant
UniProt ID: (None-None) DNA oligomer template-primer hairpin
|
|
|
|
| Sample: |
DNA polymerase E9 exo- mutant monomer, 117 kDa Vaccinia virus protein
DNA oligomer template-primer hairpin monomer, 9 kDa synthetic construct DNA
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 4 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 12
|
The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun 8(1):1455 (2017)
Tarbouriech N, Ducournau C, Hutin S, Mas PJ, Man P, Forest E, Hart DJ, Peyrefitte CN, Burmeister WP, Iseni F
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
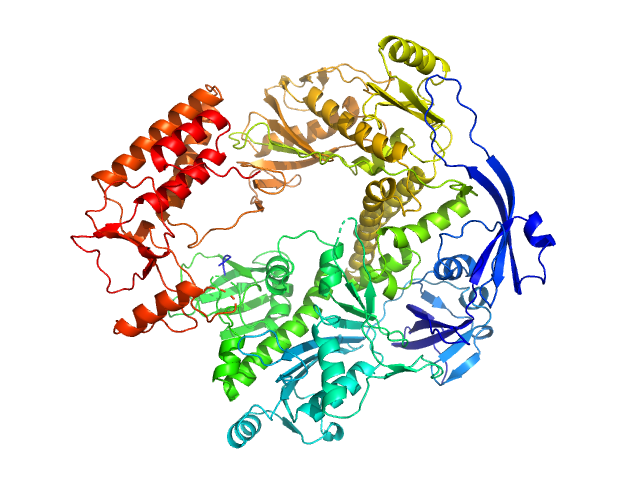
UniProt ID: P20509 (1-1006) DNA polymerase E9
|
|
|
|
| Sample: |
DNA polymerase E9 monomer, 117 kDa Vaccinia virus protein
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 4 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 27
|
The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun 8(1):1455 (2017)
Tarbouriech N, Ducournau C, Hutin S, Mas PJ, Man P, Forest E, Hart DJ, Peyrefitte CN, Burmeister WP, Iseni F
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
202 |
nm3 |
|
|
UniProt ID: P20995 (304-426) DNA polymerase processivity factor component A20 C-ter fragment
|
|
|

|
| Sample: |
DNA polymerase processivity factor component A20 C-ter fragment monomer, 17 kDa Vaccinia virus protein
|
| Buffer: |
25 mM Tris-HCl, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 5
|
The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun 8(1):1455 (2017)
Tarbouriech N, Ducournau C, Hutin S, Mas PJ, Man P, Forest E, Hart DJ, Peyrefitte CN, Burmeister WP, Iseni F
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
UniProt ID: Q25367 (1-232) S-crystallin, Q25367_DOROP
|
|
|

|
| Sample: |
S-crystallin, Q25367_DOROP dimer, 55 kDa Doryteuthis pealeii protein
|
| Buffer: |
Tissue fluid, pH: 7 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 Jul 17
|
Eye patches: Protein assembly of index-gradient squid lenses.
Science 357(6351):564-569 (2017)
Cai J, Townsend JP, Dodson TC, Heiney PA, Sweeney AM
|
|
|
UniProt ID: Q25367 (1-232) S-crystallin, Q25367_DOROP
|
|
|
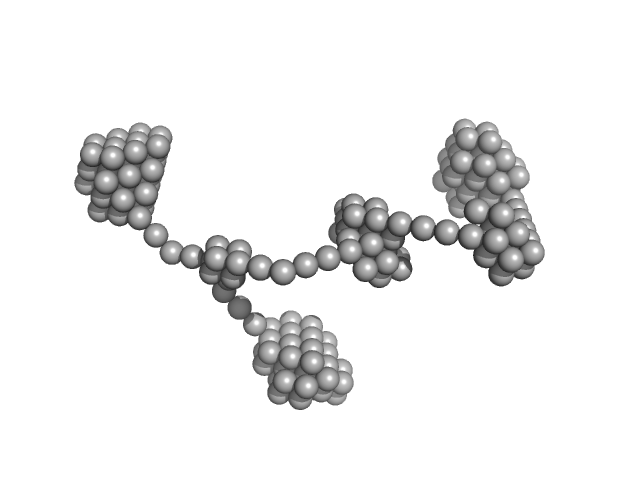
|
| Sample: |
S-crystallin, Q25367_DOROP dimer, 55 kDa Doryteuthis pealeii protein
|
| Buffer: |
Tissue fluid, pH: 7 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 Jul 19
|
Eye patches: Protein assembly of index-gradient squid lenses.
Science 357(6351):564-569 (2017)
Cai J, Townsend JP, Dodson TC, Heiney PA, Sweeney AM
|
|
|
UniProt ID: Q25367 (1-232) S-crystallin, Q25367_DOROP
|
|
|

|
| Sample: |
S-crystallin, Q25367_DOROP dimer, 55 kDa Doryteuthis pealeii protein
|
| Buffer: |
Tissue fluid, pH: 7 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 Jul 17
|
Eye patches: Protein assembly of index-gradient squid lenses.
Science 357(6351):564-569 (2017)
Cai J, Townsend JP, Dodson TC, Heiney PA, Sweeney AM
|
|
|
UniProt ID: Q25367 (1-232) S-crystallin, Q25367_DOROP
|
|
|

|
| Sample: |
S-crystallin, Q25367_DOROP dimer, 55 kDa Doryteuthis pealeii protein
|
| Buffer: |
Tissue fluid, pH: 7 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 Jul 17
|
Eye patches: Protein assembly of index-gradient squid lenses.
Science 357(6351):564-569 (2017)
Cai J, Townsend JP, Dodson TC, Heiney PA, Sweeney AM
|
|
|
UniProt ID: Q25367 (1-232) S-crystallin, Q25367_DOROP
|
|
|
|
| Sample: |
S-crystallin, Q25367_DOROP dimer, 55 kDa Doryteuthis pealeii protein
|
| Buffer: |
PBS, phosphate buffered saline, pH: 7 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2013 Nov 12
|
Eye patches: Protein assembly of index-gradient squid lenses.
Science 357(6351):564-569 (2017)
Cai J, Townsend JP, Dodson TC, Heiney PA, Sweeney AM
|
|
|
UniProt ID: A4HNB5 (None-None) Uncharacterized protein
|
|
|

|
| Sample: |
Uncharacterized protein monomer, 22 kDa Leishmania braziliensis protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM NaCl, 5 mM B-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2012 Mar 26
|
Identification of two p23 co-chaperone isoforms in Leishmania braziliensis exhibiting similar structures and Hsp90 interaction properties despite divergent stabilities.
FEBS J 282(2):388-406 (2015)
Batista FA, Almeida GS, Seraphim TV, Silva KP, Murta SM, Barbosa LR, Borges JC
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.0 |
nm |
|
|
UniProt ID: Q6PHU5 (None-None) Sortilin, also: Neurotensin-receptor 3
|
|
|

|
| Sample: |
Sortilin, also: Neurotensin-receptor 3 dimer, 153 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
275 |
nm3 |
|
|