UniProt ID: P32657 (118-1274) Chromodomain-helicase-DNA-binding protein 1
UniProt ID: (None-None) 169 bp DNA (145 bp Widom 601, flanked by 12bp DNA)
UniProt ID: P06897 (None-None) Histone H2A type 1
UniProt ID: P02281 (None-None) Histone H2B 1.1
UniProt ID: P84233 (1-136) Histone H3.2
UniProt ID: P62799 (None-None) Histone H4
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, 0.5 mM AMP-PNP, pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.6 |
nm |
| Dmax |
16.7 |
nm |
|
|
UniProt ID: P07602 (60-140) saposin-a
UniProt ID: (None-None) 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine
UniProt ID: Q97AH5 (1-197) Mechanosensitive channel T2
|
|
|

|
| Sample: |
Saposin-a monomer, 9 kDa Homo sapiens protein
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine, 47 kDa synthetic construct
Mechanosensitive channel T2 heptamer, 230 kDa Thermoplasma volcanium protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 4
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
5.4 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
686 |
nm3 |
|
|
UniProt ID: Q97AH5 (1-197) Mechanosensitive channel T2
UniProt ID: (None-None) n-Dodecyl-β-D-Maltopyranoside
|
|
|

|
| Sample: |
Mechanosensitive channel T2 heptamer, 230 kDa Thermoplasma volcanium protein
N-Dodecyl-β-D-Maltopyranoside, 77 kDa synthetic construct
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 % glycerol 0.03 % DDM, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 May 29
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
5.6 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
896 |
nm3 |
|
|
UniProt ID: P07602 (60-140) saposin-a
UniProt ID: (None-None) 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine
UniProt ID: P77304 (17-487) Dipeptide and tripeptide permease A
|
|
|

|
| Sample: |
Saposin-a monomer, 9 kDa Homo sapiens protein
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine, 47 kDa synthetic construct
Dipeptide and tripeptide permease A monomer, 51 kDa Escherichia coli protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 4
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
288 |
nm3 |
|
|
UniProt ID: P07602 (60-140) saposin-a
UniProt ID: (None-None) 1,2-dioleoyl-sn-glycero-3-phosphocholine
|
|
|

|
| Sample: |
Saposin-a monomer, 9 kDa Homo sapiens protein
1,2-dioleoyl-sn-glycero-3-phosphocholine, 47 kDa synthetic construct
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 15
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
4.1 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
UniProt ID: P07602 (60-140) saposin-a
UniProt ID: (None-None) L-α-phosphatidylinositol (soy)
|
|
|

|
| Sample: |
Saposin-a monomer, 9 kDa Homo sapiens protein
L-α-phosphatidylinositol (soy), 52 kDa Glycine max
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 15
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
3.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
UniProt ID: P07602 (60-140) saposin-a
UniProt ID: (None-None) 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol)
|
|
|

|
| Sample: |
Saposin-a monomer, 9 kDa Homo sapiens protein
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol), 46 kDa synthetic construct
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 15
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
4.1 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
UniProt ID: P33316 (94-252) Deoxyuridine 5'-triphosphate nucleotidohydrolase
|
|
|

|
| Sample: |
Deoxyuridine 5'-triphosphate nucleotidohydrolase trimer, 54 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 300 mM NaCl 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 9
|
Structural model of human dUTPase in complex with a novel proteinaceous inhibitor.
Sci Rep 8(1):4326 (2018)
Nyíri K, Mertens HDT, Tihanyi B, Nagy GN, Kőhegyi B, Matejka J, Harris MJ, Szabó JE, Papp-Kádár V, Németh-Pongrácz V, Ozohanics O, Vékey K, Svergun DI, Borysik AJ, Vértessy BG
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
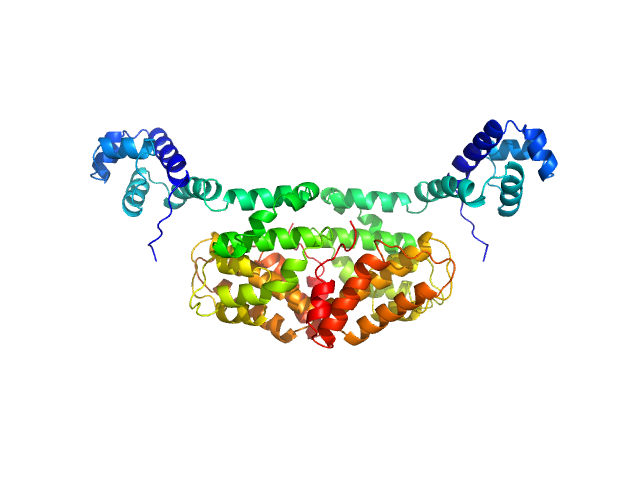
UniProt ID: E2FZP6 (13-278) SaPIbov1 pathogenicity island repressor
|
|
|
|
| Sample: |
SaPIbov1 pathogenicity island repressor dimer, 64 kDa Staphylococcus aureus protein
|
| Buffer: |
50 mM HEPES 300 mM NaCl 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 9
|
Structural model of human dUTPase in complex with a novel proteinaceous inhibitor.
Sci Rep 8(1):4326 (2018)
Nyíri K, Mertens HDT, Tihanyi B, Nagy GN, Kőhegyi B, Matejka J, Harris MJ, Szabó JE, Papp-Kádár V, Németh-Pongrácz V, Ozohanics O, Vékey K, Svergun DI, Borysik AJ, Vértessy BG
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
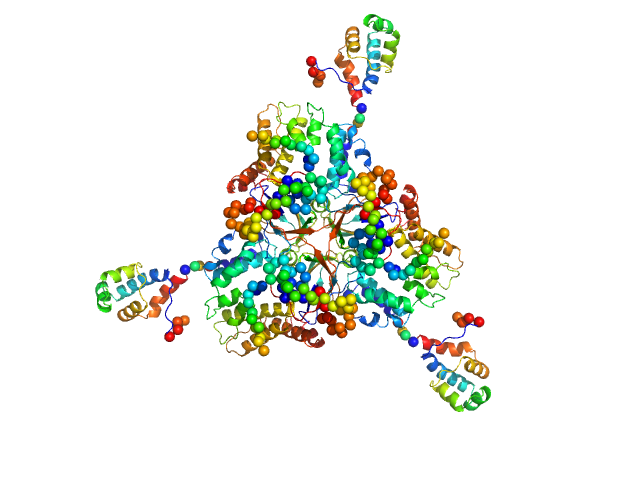
UniProt ID: P33316 (94-252) Deoxyuridine 5'-triphosphate nucleotidohydrolase
UniProt ID: E2FZP6 (13-278) SaPIbov1 pathogenicity island repressor
|
|
|
|
| Sample: |
Deoxyuridine 5'-triphosphate nucleotidohydrolase trimer, 54 kDa Homo sapiens protein
SaPIbov1 pathogenicity island repressor dimer, 64 kDa Staphylococcus aureus protein
|
| Buffer: |
50 mM HEPES 300 mM NaCl 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Structural model of human dUTPase in complex with a novel proteinaceous inhibitor.
Sci Rep 8(1):4326 (2018)
Nyíri K, Mertens HDT, Tihanyi B, Nagy GN, Kőhegyi B, Matejka J, Harris MJ, Szabó JE, Papp-Kádár V, Németh-Pongrácz V, Ozohanics O, Vékey K, Svergun DI, Borysik AJ, Vértessy BG
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
227 |
nm3 |
|
|