UniProt ID: Q5ZXN5 (19-570) Phosphocholine hydrolase Lem3
|
|
|
|
| Sample: |
Phosphocholine hydrolase Lem3 monomer, 63 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Sep 20
|
The structural analysis of dephosphocholinase Legionella pneumophila Lem3
Wenhua Zhang
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
UniProt ID: P62820-3 (None-None) Ras-related protein Rab-1A
|
|
|

|
| Sample: |
Ras-related protein Rab-1A monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Nov 17
|
The SAXS Analysis of the human GTPase Rab1a
Wenhua Zhang
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
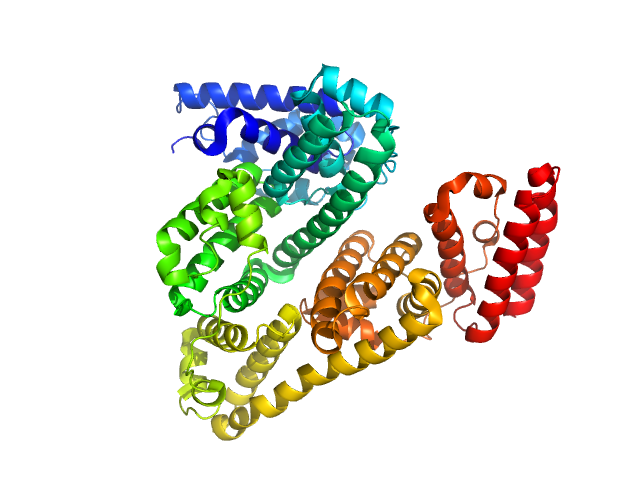
UniProt ID: P02769 (25-607) Bovine serum albumin
|
|
|
|
| Sample: |
Bovine serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
TRIS 50mM, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 21
|
A high flux setup for millisecond-scale small-angle X-ray scattering studies on macromolecular solutions
Clement Blanchet
|
|
|
UniProt ID: P24300 (None-None) Xylose isomerase
|
|
|
|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
100 mM HEPES, 1 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 21
|
A high flux setup for millisecond-scale small-angle X-ray scattering studies on macromolecular solutions
Clement Blanchet
|
|
|
UniProt ID: P62894 (None-None) Cytochrome C
|
|
|
|
| Sample: |
Cytochrome C monomer, 12 kDa Bos taurus protein
|
| Buffer: |
TRIS 50mM, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 21
|
A high flux setup for millisecond-scale small-angle X-ray scattering studies on macromolecular solutions
Clement Blanchet
|
|
|
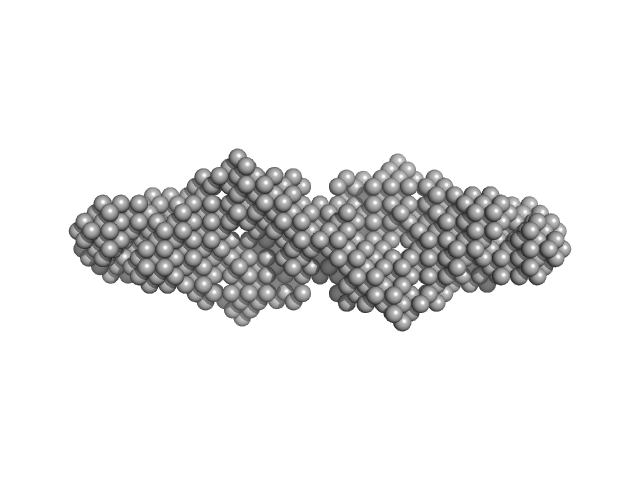
UniProt ID: G0SFY0 (1-136) hypothetical protein CTHT_0072540
|
|
|
|
| Sample: |
Hypothetical protein CTHT_0072540 tetramer, 62 kDa Chaetomium thermophilum protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 2 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 30
|
Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol Cell 69(6):979-992.e6 (2018)
de Moura TR, Mozaffari-Jovin S, Szabó CZK, Schmitzová J, Dybkov O, Cretu C, Kachala M, Svergun D, Urlaub H, Lührmann R, Pena V
|
| RgGuinier |
4.1 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
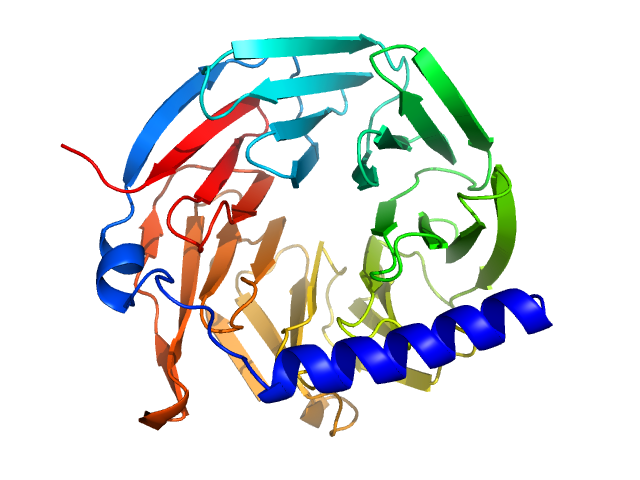
UniProt ID: G0SFY0 (155-480) hypothetical protein CTHT_0072540
|
|
|
|
| Sample: |
Hypothetical protein CTHT_0072540 monomer, 35 kDa Chaetomium thermophilum protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 2 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 19
|
Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol Cell 69(6):979-992.e6 (2018)
de Moura TR, Mozaffari-Jovin S, Szabó CZK, Schmitzová J, Dybkov O, Cretu C, Kachala M, Svergun D, Urlaub H, Lührmann R, Pena V
|
| RgGuinier |
2.3 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
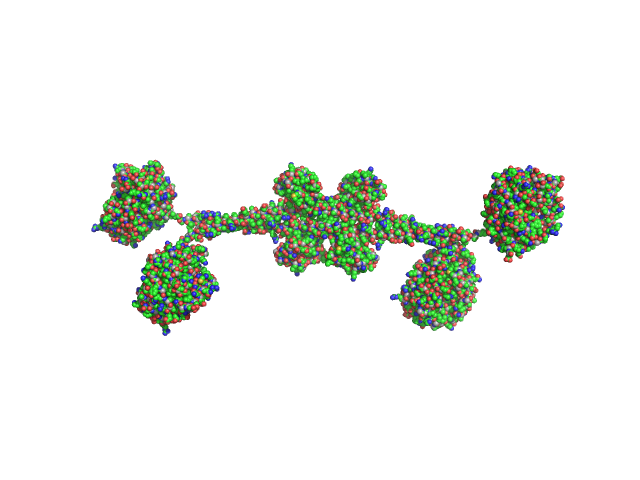
UniProt ID: G0SFY0 (1-480) Full-length hypothetical protein CTHT_0072540
|
|
|
|
| Sample: |
Full-length hypothetical protein CTHT_0072540 tetramer, 207 kDa Chaetomium thermophilum protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 2 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2013 Oct 15
|
Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol Cell 69(6):979-992.e6 (2018)
de Moura TR, Mozaffari-Jovin S, Szabó CZK, Schmitzová J, Dybkov O, Cretu C, Kachala M, Svergun D, Urlaub H, Lührmann R, Pena V
|
| RgGuinier |
6.2 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
UniProt ID: Q9UI95 (1-211) Mitotic spindle assembly checkpoint protein MAD2B
UniProt ID: O60673 (1988-2014) DNA polymerase zeta catalytic subunit
|
|
|
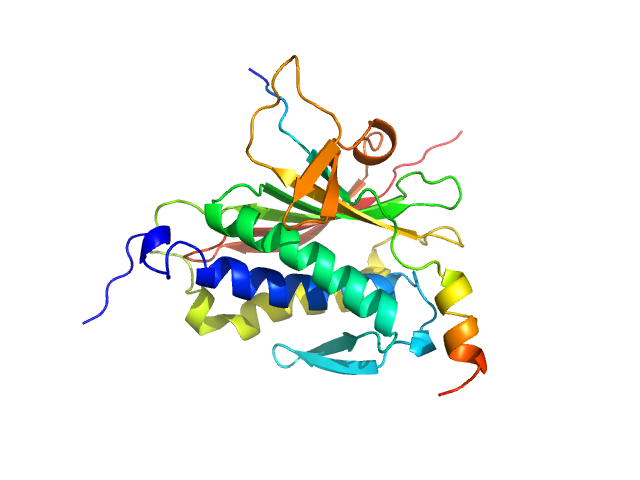
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B monomer, 24 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol, pH: 8.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: Q9UI95 (1-211) Mitotic spindle assembly checkpoint protein MAD2B
UniProt ID: O60673 (1988-2014) DNA polymerase zeta catalytic subunit
|
|
|
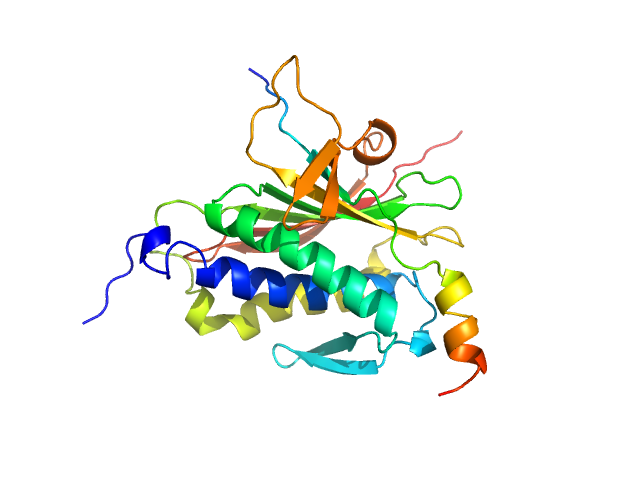
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B monomer, 24 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol, pH: 8.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
47 |
nm3 |
|
|