UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase, reductase component
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase, reductase component dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 5% glycerol, 1 mM HPA, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Mar 7
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase, reductase component
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase, reductase component dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 5% glycerol, 1 mM HPA, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Mar 7
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase, reductase component
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase, reductase component dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 5% glycerol, 1 mM HPA, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Mar 7
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
UniProt ID: P53776 (24-149) LIM/homeobox protein Lhx4
UniProt ID: Q9CXV0 (273-301) Insulin gene enhancer protein ISL-2
|
|
|
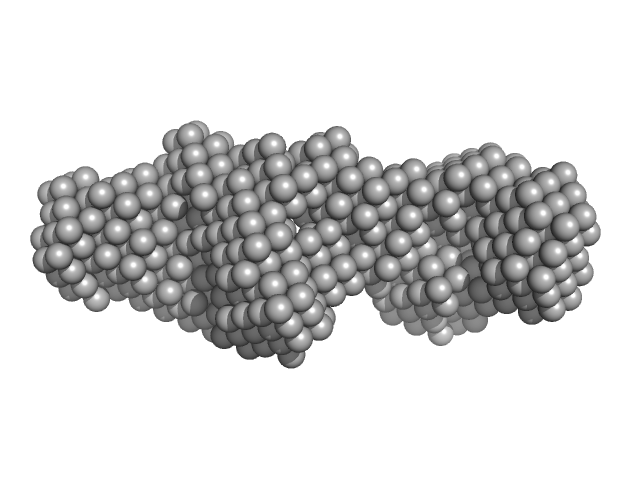
|
| Sample: |
LIM/homeobox protein Lhx4 monomer, 15 kDa Mus musculus protein
Insulin gene enhancer protein ISL-2 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins (2019)
Stokes PH, Robertson NO, Silva AP, Estephan T, Trewhella J, Guss JM, Matthews JM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
UniProt ID: P53776 (24-149) LIM/homeobox protein Lhx4
UniProt ID: Q9CXV0 (273-301) Insulin gene enhancer protein ISL-2 (R282G)
|
|
|
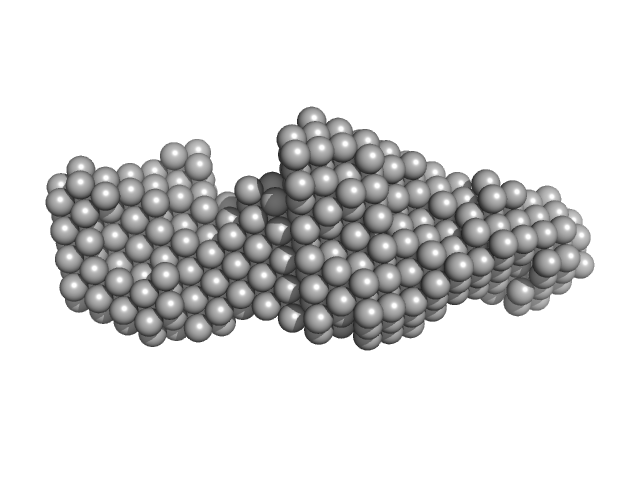
|
| Sample: |
LIM/homeobox protein Lhx4 monomer, 15 kDa Mus musculus protein
Insulin gene enhancer protein ISL-2 (R282G) monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins (2019)
Stokes PH, Robertson NO, Silva AP, Estephan T, Trewhella J, Guss JM, Matthews JM
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
UniProt ID: P00516 (54-671) cGMP-dependent protein kinase 1
|
|
|

|
| Sample: |
CGMP-dependent protein kinase 1 monomer, 70 kDa Bos taurus protein
|
| Buffer: |
50 mM MES, 300 mM NaCl, 1 mM TCEP, 5 mM DTT, pH: 6.9 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 Jun 6
|
An N-terminally truncated form of cyclic GMP-dependent protein kinase Iα (PKG Iα) is monomeric and autoinhibited and provides a model for activation.
J Biol Chem 293(21):7916-7929 (2018)
Moon TM, Sheehe JL, Nukareddy P, Nausch LW, Wohlfahrt J, Matthews DE, Blumenthal DK, Dostmann WR
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
UniProt ID: P01857 (None-None) Immunoglobulin heavy constant gamma 1
|
|
|
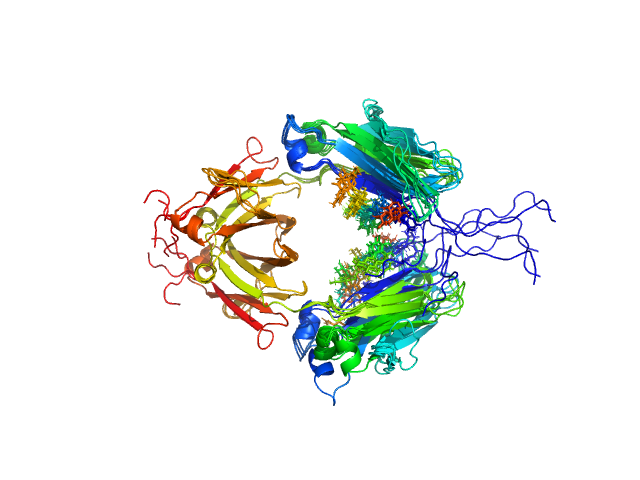
|
| Sample: |
Immunoglobulin heavy constant gamma 1 dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: P01859 (None-None) Immunoglobulin heavy constant gamma 2
|
|
|
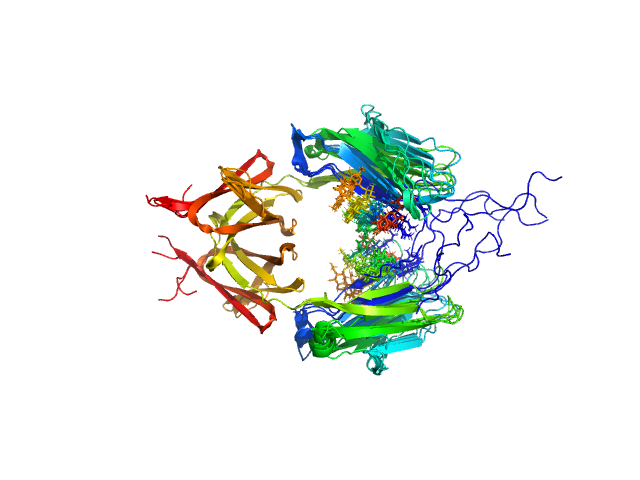
|
| Sample: |
Immunoglobulin heavy constant gamma 2 dimer, 52 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
UniProt ID: P01857 (None-None) Immunoglobulin heavy constant gamma 1 M255Y/S257T/T259E
|
|
|
|
| Sample: |
Immunoglobulin heavy constant gamma 1 M255Y/S257T/T259E dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
UniProt ID: O60568 (25-738) Procollagen lysyl hydroxylase LH3
|
|
|

|
| Sample: |
Procollagen lysyl hydroxylase LH3 dimer, 180 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 28
|
Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun 9(1):3163 (2018)
Scietti L, Chiapparino A, De Giorgi F, Fumagalli M, Khoriauli L, Nergadze S, Basu S, Olieric V, Cucca L, Banushi B, Profumo A, Giulotto E, Gissen P, Forneris F
|
| RgGuinier |
5.1 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
268 |
nm3 |
|
|