UniProt ID: P19339 (123-294) Protein sex-lethal
|
|
|
|
| Sample: |
Protein sex-lethal monomer, 20 kDa Drosophila melanogaster protein
|
| Buffer: |
10 mM KP, 50 mM NaCl, 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 16
|
A General Small-Angle X-ray Scattering-Based Screening Protocol Validated for Protein-RNA Interactions.
ACS Comb Sci 20(4):197-202 (2018)
Chen PC, Masiewicz P, Rybin V, Svergun D, Hennig J
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
UniProt ID: P19339 (123-294) Protein sex-lethal mutant
|
|
|
|
| Sample: |
Protein sex-lethal mutant monomer, 20 kDa Drosophila melanogaster protein
|
| Buffer: |
10 mM KP, 50 mM NaCl, 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 2
|
A General Small-Angle X-ray Scattering-Based Screening Protocol Validated for Protein-RNA Interactions.
ACS Comb Sci 20(4):197-202 (2018)
Chen PC, Masiewicz P, Rybin V, Svergun D, Hennig J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: P19339 (123-294) Protein sex-lethal mutant
UniProt ID: None (None-None) RNA decaneucleotide U8GU
|
|
|
|
| Sample: |
Protein sex-lethal mutant monomer, 20 kDa Drosophila melanogaster protein
RNA decaneucleotide U8GU monomer, 3 kDa synthetic construct RNA
|
| Buffer: |
50% dilution of protein buffer {10 mM KP, 50 mM NaCl, 10 mM DTT pH 6} with {milliQ-water pH 7} suspended RNA, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 3
|
A General Small-Angle X-ray Scattering-Based Screening Protocol Validated for Protein-RNA Interactions.
ACS Comb Sci 20(4):197-202 (2018)
Chen PC, Masiewicz P, Rybin V, Svergun D, Hennig J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
UniProt ID: P19339 (123-294) Protein sex-lethal mutant
UniProt ID: None (None-None) RNA decaneucleotide UGU8
|
|
|
|
| Sample: |
Protein sex-lethal mutant dimer, 41 kDa Drosophila melanogaster protein
RNA decaneucleotide UGU8 dimer, 6 kDa synthetic construct RNA
|
| Buffer: |
50% dilution of protein buffer {10 mM KP, 50 mM NaCl, 10 mM DTT pH 6} with {milliQ-water pH 7} suspended RNA, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 12
|
A General Small-Angle X-ray Scattering-Based Screening Protocol Validated for Protein-RNA Interactions.
ACS Comb Sci 20(4):197-202 (2018)
Chen PC, Masiewicz P, Rybin V, Svergun D, Hennig J
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
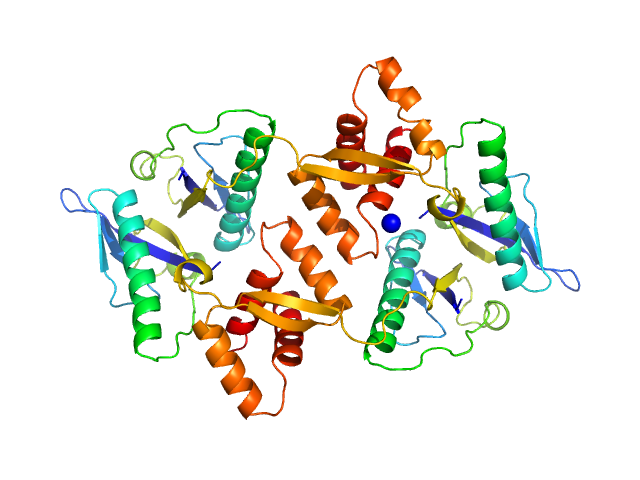
UniProt ID: Q63NA5 (1-134) Type II toxin-antitoxin system HicB family antitoxin
|
|
|
|
| Sample: |
Type II toxin-antitoxin system HicB family antitoxin tetramer, 63 kDa Burkholderia pseudomallei protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 28
|
The molecular basis of protein toxin HicA-dependent binding of the protein antitoxin HicB to DNA.
J Biol Chem (2018)
Winter AJ, Williams C, Isupov MN, Crocker H, Gromova M, Marsh P, Wilkinson OJ, Dillingham MS, Harmer NJ, Titball RW, Crump MP
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
112 |
nm3 |
|
|
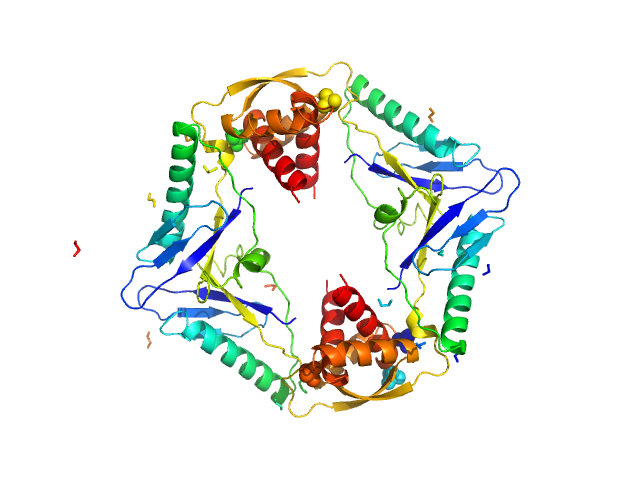
UniProt ID: Q63NA5 (1-134) Type II toxin-antitoxin system HicB family antitoxin
UniProt ID: Q63NA6 (None-None) Addiction module toxin, HicA
|
|
|
|
| Sample: |
Type II toxin-antitoxin system HicB family antitoxin tetramer, 63 kDa Burkholderia pseudomallei protein
Addiction module toxin, HicA monomer, 7 kDa Burkholderia pseudomallei protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 27
|
The molecular basis of protein toxin HicA-dependent binding of the protein antitoxin HicB to DNA.
J Biol Chem (2018)
Winter AJ, Williams C, Isupov MN, Crocker H, Gromova M, Marsh P, Wilkinson OJ, Dillingham MS, Harmer NJ, Titball RW, Crump MP
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
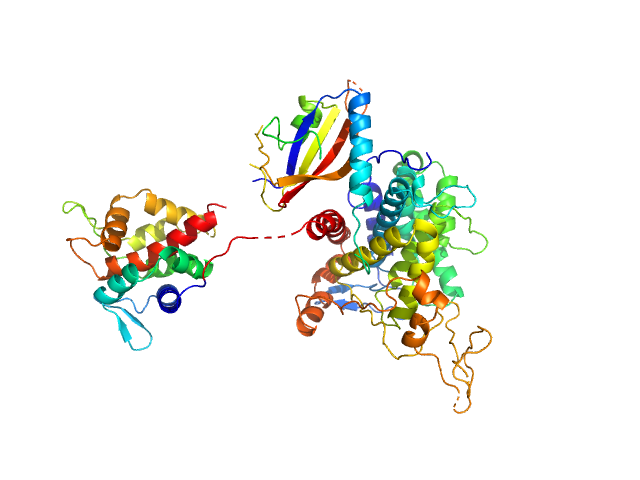
UniProt ID: Q5ZTK4 (277-907) Ubiquitinating/deubiquitinating enzyme SdeA
|
|
|
|
| Sample: |
Ubiquitinating/deubiquitinating enzyme SdeA monomer, 72 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
10 mM HEPES 150 mM NaCl 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 26
|
Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination.
Nature 557(7707):734-738 (2018)
Kalayil S, Bhogaraju S, Bonn F, Shin D, Liu Y, Gan N, Basquin J, Grumati P, Luo ZQ, Dikic I
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
UniProt ID: Q15583 (151-248) Homeobox protein TGIF1
|
|
|
|
| Sample: |
Homeobox protein TGIF1 dimer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 18
|
'TG' interacting factor
Ewelina Guca
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
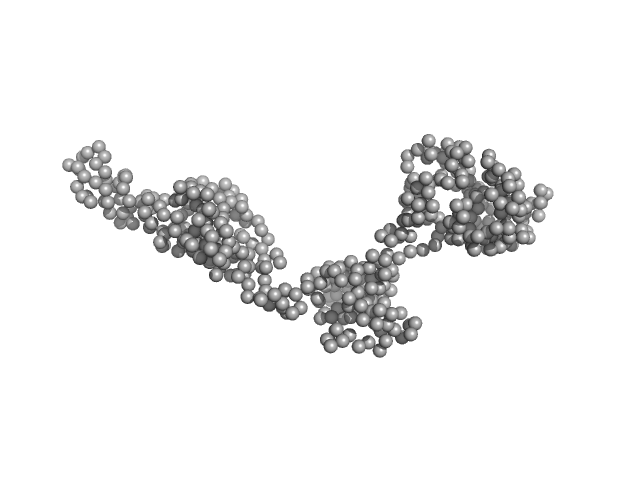
UniProt ID: Q5T2W7 (7-460) Uncharacterized protein C1orf159
|
|
|
|
| Sample: |
Uncharacterized protein C1orf159 monomer, 53 kDa Homo sapiens protein
|
| Buffer: |
20mM Hepes 150 NaCl 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 28
|
Probing the Architecture of a Multi-PDZ Domain Protein: Structure of PDZK1 in Solution.
Structure 26(11):1522-1533.e5 (2018)
Hajizadeh NR, Pieprzyk J, Skopintsev P, Flayhan A, Svergun DI, Löw C
|
| RgGuinier |
3.9 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
UniProt ID: Q9ULB1 (904-1307) Neurexin 1a L5L6
|
|
|
|
| Sample: |
Neurexin 1a L5L6 monomer, 44 kDa protein
|
| Buffer: |
20 mM HEPES pH 8, 150 mM NaCl, 0.5mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2016 Sep 6
|
Structural Plasticity of Neurexin 1α: Implications for its Role as Synaptic Organizer.
J Mol Biol 430(21):4325-4343 (2018)
Liu J, Misra A, Reddy MVVVS, White MA, Ren G, Rudenko G
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
69 |
nm3 |
|
|