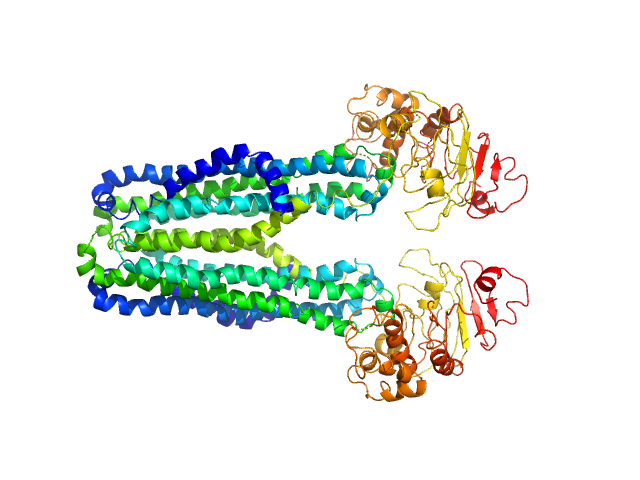
UniProt ID: Q9ULB1 (904-1307) Neurexin 1a L5L6 with ss6 insert
|
|
|
|
| Sample: |
Neurexin 1a L5L6 with ss6 insert monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 8, 150 mM NaCl, 0.5mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2016 Sep 26
|
Structural Plasticity of Neurexin 1α: Implications for its Role as Synaptic Organizer.
J Mol Biol 430(21):4325-4343 (2018)
Liu J, Misra A, Reddy MVVVS, White MA, Ren G, Rudenko G
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: P60752 (None-None) Lipid A export ATP-binding/permease protein MsbA
UniProt ID: None (None-None) Membrane scaffold protein 1D1 (deuterated, 75%)
UniProt ID: None (None-None) 1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl)
|
|
|
|
| Sample: |
Lipid A export ATP-binding/permease protein MsbA dimer, 133 kDa Escherichia coli protein
Membrane scaffold protein 1D1 (deuterated, 75%) dimer, 49 kDa protein
1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), 1 kDa Escherichia coli
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at D11, ILL on 2017 Mar 9
|
Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs.
Structure 26(8):1072-1079.e4 (2018)
Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, Tidow H
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
UniProt ID: P60752 (None-None) Lipid A export ATP-binding/permease protein MsbA
UniProt ID: None (None-None) Membrane scaffold protein 1D1 (deuterated, 75%)
UniProt ID: None (None-None) 1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl)
|
|
|
|
| Sample: |
Lipid A export ATP-binding/permease protein MsbA dimer, 133 kDa Escherichia coli protein
Membrane scaffold protein 1D1 (deuterated, 75%) dimer, 49 kDa protein
1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), 1 kDa Escherichia coli
|
| Buffer: |
30 mM Tris, 150 mM NaCl, 1 mM ADP, pH: 7.5 |
| Experiment: |
SANS
data collected at D11, ILL on 2017 Mar 9
|
Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs.
Structure 26(8):1072-1079.e4 (2018)
Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, Tidow H
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
173 |
nm3 |
|
|
UniProt ID: P60752 (None-None) Lipid A export ATP-binding/permease protein MsbA
UniProt ID: None (None-None) Membrane scaffold protein 1D1 (deuterated, 75%)
UniProt ID: None (None-None) 1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl)
|
|
|
|
| Sample: |
Lipid A export ATP-binding/permease protein MsbA dimer, 133 kDa Escherichia coli protein
Membrane scaffold protein 1D1 (deuterated, 75%) dimer, 49 kDa protein
1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), 1 kDa Escherichia coli
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 8
|
Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs.
Structure 26(8):1072-1079.e4 (2018)
Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, Tidow H
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
607 |
nm3 |
|
|
UniProt ID: P60752 (None-None) Nucleotide Binding Domain of Lipid A export ATP-binding/permease protein MsbA
|
|
|
|
| Sample: |
Nucleotide Binding Domain of Lipid A export ATP-binding/permease protein MsbA monomer, 27 kDa Escherichia coli protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 30
|
Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs.
Structure 26(8):1072-1079.e4 (2018)
Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, Tidow H
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
UniProt ID: P60752 (None-None) Nucleotide Binding Domain of Lipid A export ATP-binding/permease protein MsbA
|
|
|
|
| Sample: |
Nucleotide Binding Domain of Lipid A export ATP-binding/permease protein MsbA monomer, 27 kDa Escherichia coli protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, 0.5 mM TCEP, 1 mM ADP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 30
|
Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs.
Structure 26(8):1072-1079.e4 (2018)
Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, Tidow H
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
UniProt ID: Q9DH28 (None-None) Nonstructural protein sigma NS
UniProt ID: None (None-None) 20mer RNA (unstructured)
|
|
|
|
| Sample: |
Nonstructural protein sigma NS octamer, 325 kDa Avian orthoreovirus protein
20mer RNA (unstructured) dimer, 13 kDa RNA
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 25
|
Stability of local secondary structure determines selectivity of viral RNA chaperones.
Nucleic Acids Res (2018)
Bravo JPK, Borodavka A, Barth A, Calabrese AN, Mojzes P, Cockburn JJB, Lamb DC, Tuma R
|
| RgGuinier |
7.8 |
nm |
| Dmax |
38.0 |
nm |
| VolumePorod |
964 |
nm3 |
|
|
UniProt ID: Q9DH28 (None-None) Nonstructural protein sigma NS
|
|
|
|
| Sample: |
Nonstructural protein sigma NS hexamer, 244 kDa Avian orthoreovirus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 25
|
Stability of local secondary structure determines selectivity of viral RNA chaperones.
Nucleic Acids Res (2018)
Bravo JPK, Borodavka A, Barth A, Calabrese AN, Mojzes P, Cockburn JJB, Lamb DC, Tuma R
|
| RgGuinier |
5.5 |
nm |
| Dmax |
23.1 |
nm |
| VolumePorod |
670 |
nm3 |
|
|
UniProt ID: A5I6N2 (2-292) 4-hydroxy-tetrahydrodipicolinate synthase from Clostridium botulinum
|
|
|
|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase from Clostridium botulinum tetramer, 126 kDa Clostridium botulinum protein
|
| Buffer: |
20mM Tris, 150mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2010 Nov 26
|
Substrate Locking Promotes Dimer-Dimer Docking of an Enzyme Antibiotic Target.
Structure 26(7):948-959.e5 (2018)
Atkinson SC, Dogovski C, Wood K, Griffin MDW, Gorman MA, Hor L, Reboul CF, Buckle AM, Wuttke J, Parker MW, Dobson RCJ, Perugini MA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
159 |
nm3 |
|
|
UniProt ID: A5I6N2 (2-292) 4-hydroxy-tetrahydrodipicolinate synthase from Clostridium botulinum
|
|
|
|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase from Clostridium botulinum tetramer, 126 kDa Clostridium botulinum protein
|
| Buffer: |
20mM Tris, 150mM NaCl, 5mM sodium pyruvate, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2010 Nov 26
|
Substrate Locking Promotes Dimer-Dimer Docking of an Enzyme Antibiotic Target.
Structure 26(7):948-959.e5 (2018)
Atkinson SC, Dogovski C, Wood K, Griffin MDW, Gorman MA, Hor L, Reboul CF, Buckle AM, Wuttke J, Parker MW, Dobson RCJ, Perugini MA
|
| RgGuinier |
3.3 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
165 |
nm3 |
|
|