|
|
|
|
|
| Sample: |
Gamma Cyclodextrin monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at 11-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 2
|
Extended q‐range X‐ray Scattering Reveals High‐Resolution Structural Details of Biomacromolecules in Aqueous Solutions
Chemistry – A European Journal (2023)
Chen J, Borkiewicz O, Grishaev A, Zhang F, Bera M, Ruett U, Levin I
|
|
|
|
|
|
|
|
| Sample: |
Gamma Cyclodextrin monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at 11-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 2
|
Extended q‐range X‐ray Scattering Reveals High‐Resolution Structural Details of Biomacromolecules in Aqueous Solutions
Chemistry – A European Journal (2023)
Chen J, Borkiewicz O, Grishaev A, Zhang F, Bera M, Ruett U, Levin I
|
|
|
|
|
|
|
|
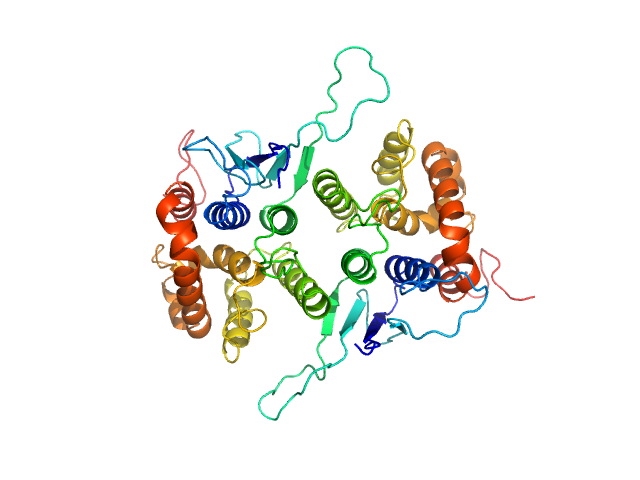
| Sample: |
His-tagged fused complex of cytochrome P450 143 and ferredoxin Rv1786 monomer, 54 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
300 mM NaCl, 50 mM Tris/TrisHCl, 10% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 7
|
Structural insights into 3Fe–4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450
Frontiers in Molecular Biosciences 9 (2023)
Gilep A, Varaksa T, Bukhdruker S, Kavaleuski A, Ryzhykau Y, Smolskaya S, Sushko T, Tsumoto K, Grabovec I, Kapranov I, Okhrimenko I, Marin E, Shevtsov M, Mishin A, Kovalev K, Kuklin A, Gordeliy V, Kaluzhskiy L, Gnedenko O, Yablokov E, Ivanov A, Borshchevskiy V, Strushkevich N
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glutamate--tRNA ligase dimer, 57 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
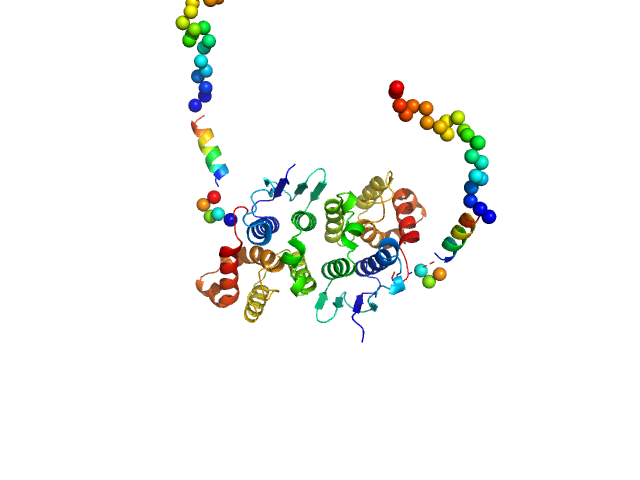
|
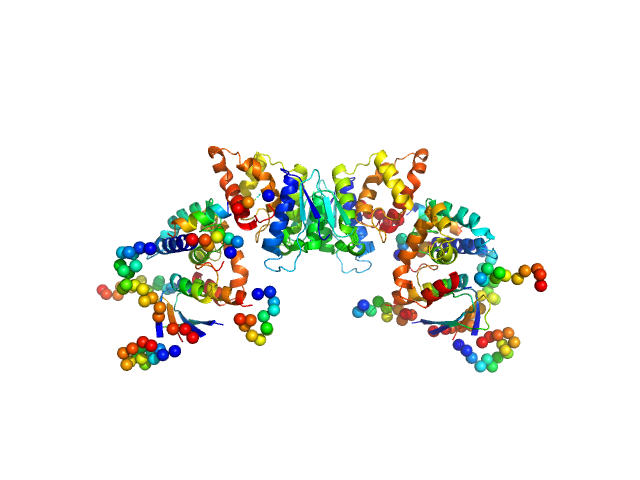
| Sample: |
TRNA import protein tRIP dimer, 49 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
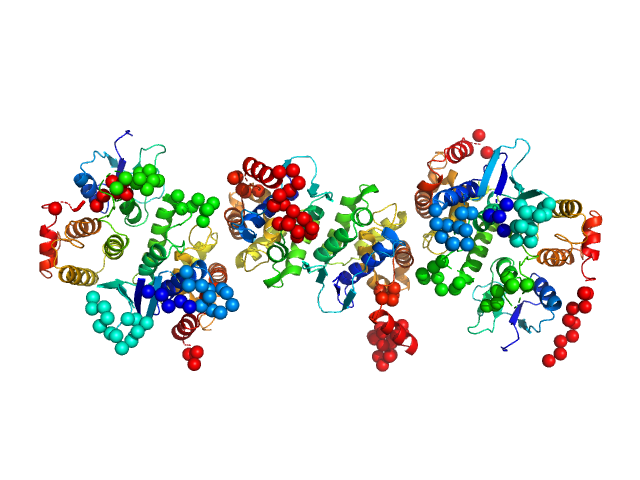
| Sample: |
Chimeric construct containing the GST-like domains of P. berghei tRNA import protein and glutamyl-tRNA synthetase dimer, 105 kDa Plasmodium berghei ANKA protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
|
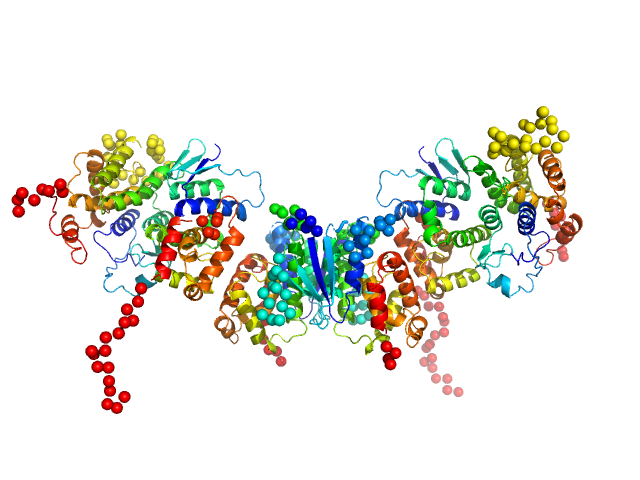
| Sample: |
Glutamine--tRNA ligase dimer, 46 kDa Plasmodium berghei (strain … protein
Glutamate--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
TRNA import protein tRIP dimer, 48 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
198 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Methionine--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
TRNA import protein tRIP dimer, 48 kDa Plasmodium berghei (strain … protein
Glutamate--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|
|
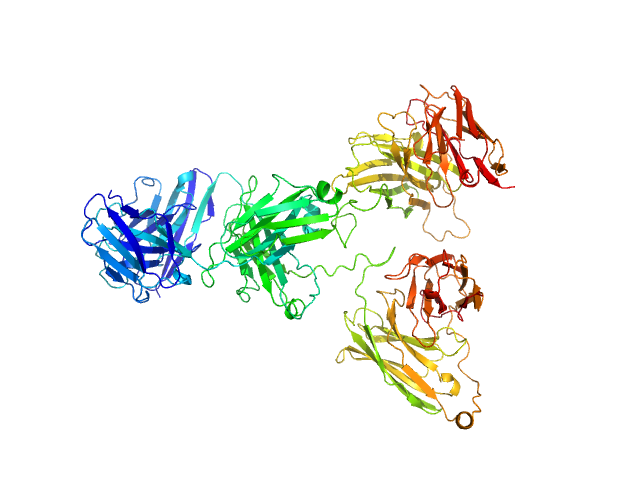
| Sample: |
Tri-specific antibody monomer, 102 kDa Homo sapiens protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Apr 26
|
TRYBE®: an Fc-free antibody format with three monovalent targeting arms engineered for long in vivo half-life.
MAbs 15(1):2160229 (2023)
Davé E, Durrant O, Dhami N, Compson J, Broadbridge J, Archer S, Maroof A, Whale K, Menochet K, Bonnaillie P, Barry E, Wild G, Peerboom C, Bhatta P, Ellis M, Hinchliffe M, Humphreys DP, Heywood SP
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
High affinity immunoglobulin gamma Fc recombinant CD64 monomer, 32 kDa synthetic construct protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Dec 9
|
The solution structure of the unbound IgG Fc receptor CD64 resembles its crystal structure: Implications for function.
PLoS One 18(9):e0288351 (2023)
Hui GK, Gao X, Gor J, Lu J, Sun PD, Perkins SJ
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
30 |
nm3 |
|
|