|
|
|
|
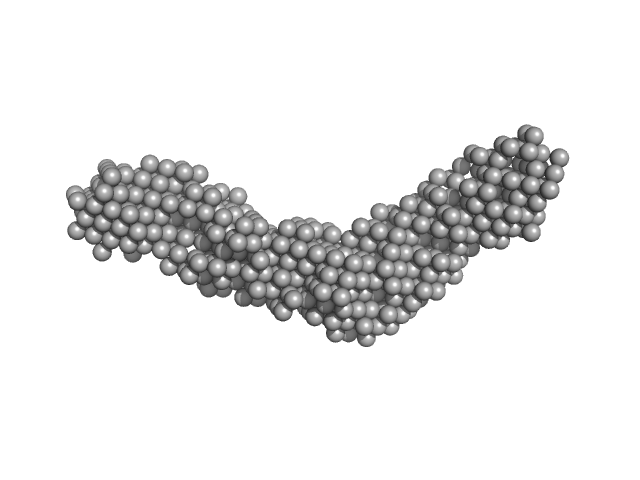
|
| Sample: |
ColH protein monomer, 34 kDa Hathewaya histolytica protein
Collagenous Peptide model [(PPG)10] trimer, 10 kDa synthetic construct protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ -induced orientation of tandem collagen binding domains from clostridial collagenase ColG permits two opposing functions of collagen fibril formation and retardation.
FEBS J 285(17):3254-3269 (2018)
Caviness P, Bauer R, Tanaka K, Janowska K, Roeser JR, Harter D, Sanders J, Ruth C, Matsushita O, Sakon J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
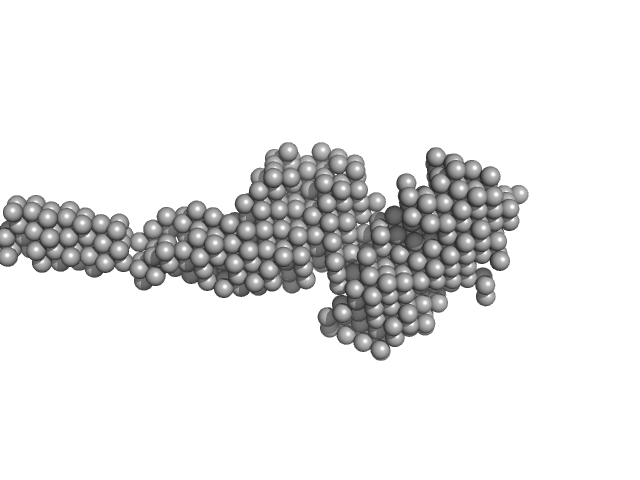
|
| Sample: |
Collagenous Peptide model [(PPG)10] trimer, 9 kDa synthetic construct protein
ColG Collagenase monomer, 37 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ -induced orientation of tandem collagen binding domains from clostridial collagenase ColG permits two opposing functions of collagen fibril formation and retardation.
FEBS J 285(17):3254-3269 (2018)
Caviness P, Bauer R, Tanaka K, Janowska K, Roeser JR, Harter D, Sanders J, Ruth C, Matsushita O, Sakon J
|
| RgGuinier |
4.1 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
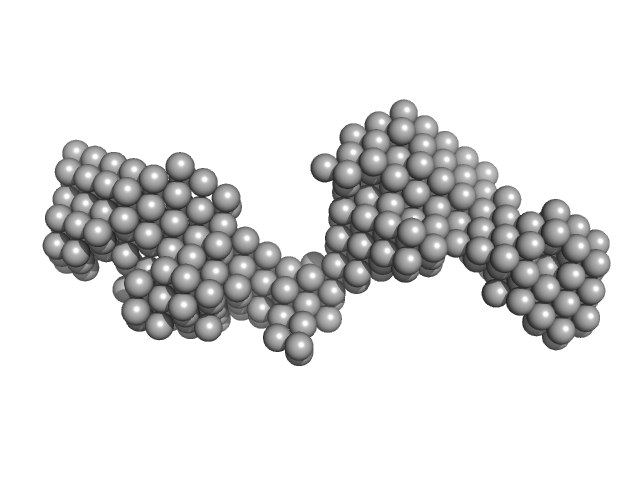
|
| Sample: |
Tegument protein UL37 monomer, 49 kDa Suid alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Inner tegument protein monomer, 62 kDa Human alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
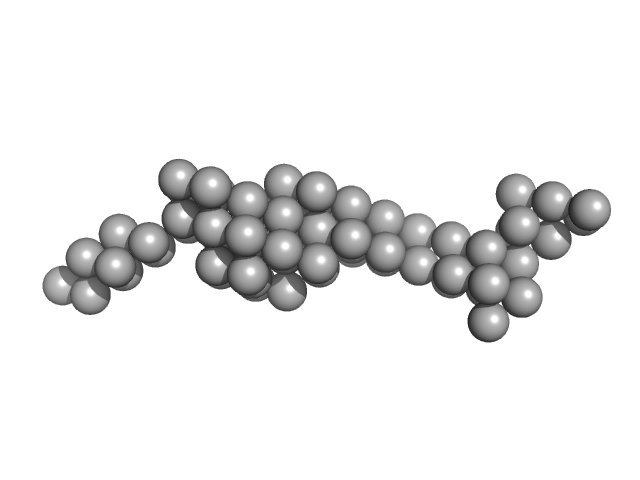
|
| Sample: |
Tegument protein UL37 monomer, 43 kDa Suid alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
|
|
|
|
|
|
|
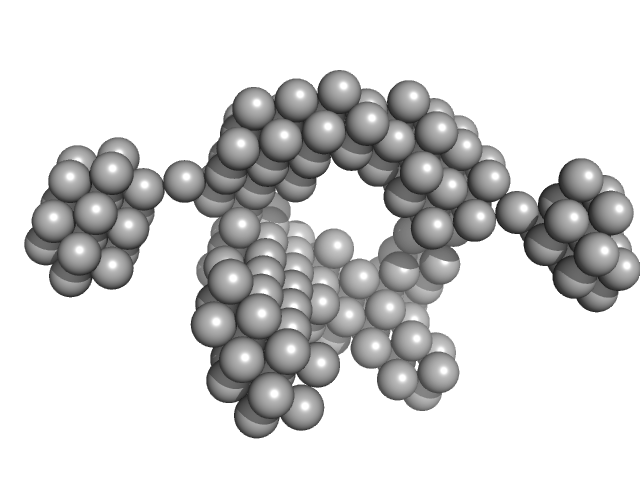
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M NaCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
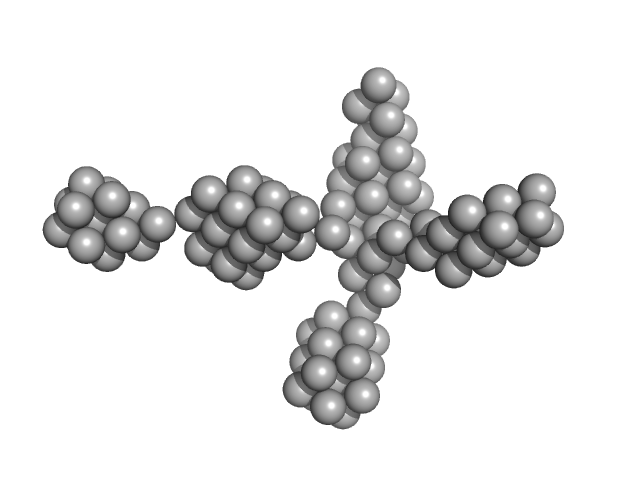
|
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M KBr, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
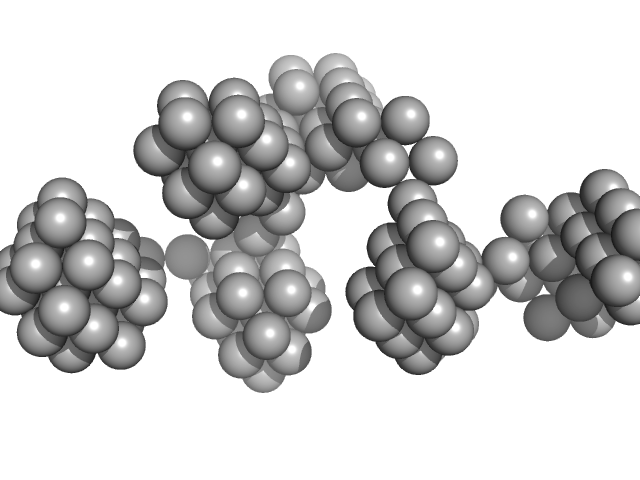
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M NaBr, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 26
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
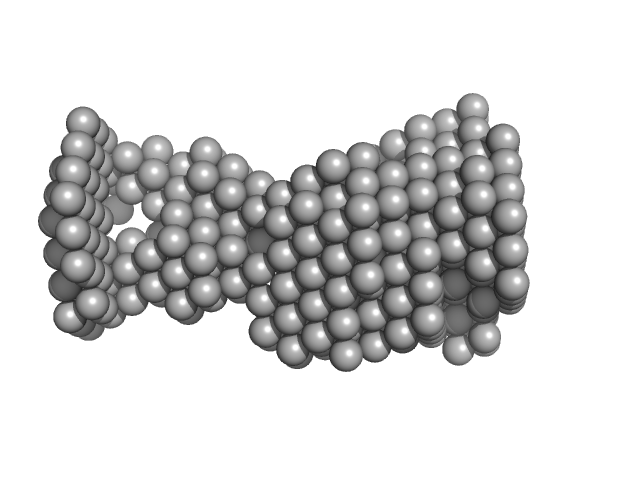
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M RbCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Procollagen lysyl hydroxylase LH3 dimer, 180 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 28
|
Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun 9(1):3163 (2018)
Scietti L, Chiapparino A, De Giorgi F, Fumagalli M, Khoriauli L, Nergadze S, Basu S, Olieric V, Cucca L, Banushi B, Profumo A, Giulotto E, Gissen P, Forneris F
|
| RgGuinier |
5.1 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
268 |
nm3 |
|
|