|
|
|
|
|
| Sample: |
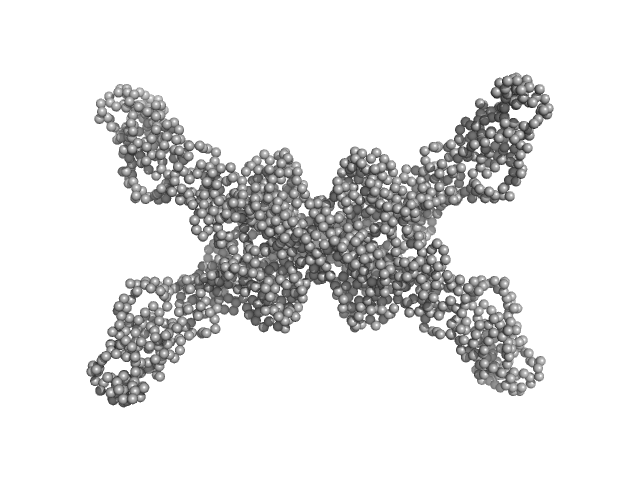
Geobacillus stearothermophilus DnaB full-length tetramer, 214 kDa Geobacillus stearothermophilus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl and 5 mM β-ME, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 Mar 12
|
Structural analyses of the bacterial primosomal protein DnaB reveal that it is a tetramer and forms a complex with a primosomal re-initiation protein.
J Biol Chem 292(38):15744-15757 (2017)
Li YC, Naveen V, Lin MG, Hsiao CD
|
| RgGuinier |
5.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
432 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peptidylprolyl isomerase monomer, 54 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
20 mM Tris pH 7.5, 150 mM KCl, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 30
|
Structural and molecular comparison of bacterial and eukaryotic trigger factors.
Sci Rep 7(1):10680 (2017)
Ries F, Carius Y, Rohr M, Gries K, Keller S, Lancaster CRD, Willmund F
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Trigger factor-like protein TIG, Chloroplastic monomer, 53 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris pH 7.5, 150 mM KCl, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 30
|
Structural and molecular comparison of bacterial and eukaryotic trigger factors.
Sci Rep 7(1):10680 (2017)
Ries F, Carius Y, Rohr M, Gries K, Keller S, Lancaster CRD, Willmund F
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
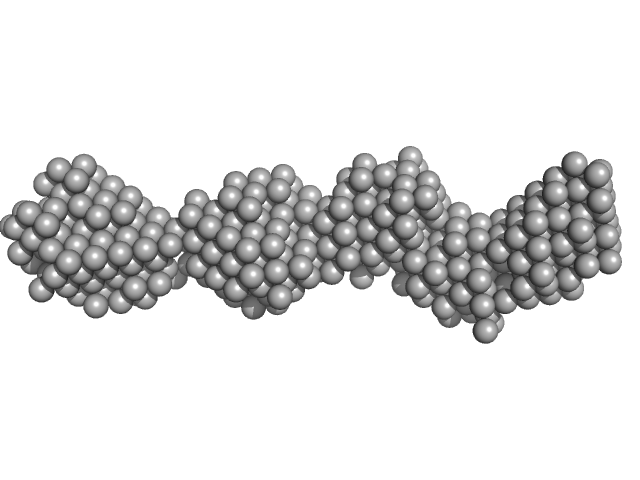
40bp long dsDNA-Sa Oligonucleotide monomer, 25 kDa DNA
|
| Buffer: |
0.5 x Tris/Borate/EDTA (TBE), pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 28
|
Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius.
Mol Microbiol 105(5):777-793 (2017)
Li L, Banerjee A, Bischof LF, Maklad HR, Hoffmann L, Henche AL, Veliz F, Bildl W, Schulte U, Orell A, Essen LO, Peeters E, Albers SV
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Wild-type archaeal biofilm regulator 1 (ABfR1: Transcriptional regulator ArsR family). dimer, 26 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 27
|
Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius.
Mol Microbiol 105(5):777-793 (2017)
Li L, Banerjee A, Bischof LF, Maklad HR, Hoffmann L, Henche AL, Veliz F, Bildl W, Schulte U, Orell A, Essen LO, Peeters E, Albers SV
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Archaeal biofilm regulator 1 (AbfR1) mutant Y84E S87D phosphomimic mutant dimer, 26 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 27
|
Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius.
Mol Microbiol 105(5):777-793 (2017)
Li L, Banerjee A, Bischof LF, Maklad HR, Hoffmann L, Henche AL, Veliz F, Bildl W, Schulte U, Orell A, Essen LO, Peeters E, Albers SV
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
40bp long dsDNA-Sa Oligonucleotide monomer, 25 kDa DNA
Wild-type archaeal biofilm regulator 1 (ABfR1: Transcriptional regulator ArsR family). dimer, 26 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
0.5 x Tris/Borate/EDTA (TBE), pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 27
|
Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius.
Mol Microbiol 105(5):777-793 (2017)
Li L, Banerjee A, Bischof LF, Maklad HR, Hoffmann L, Henche AL, Veliz F, Bildl W, Schulte U, Orell A, Essen LO, Peeters E, Albers SV
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
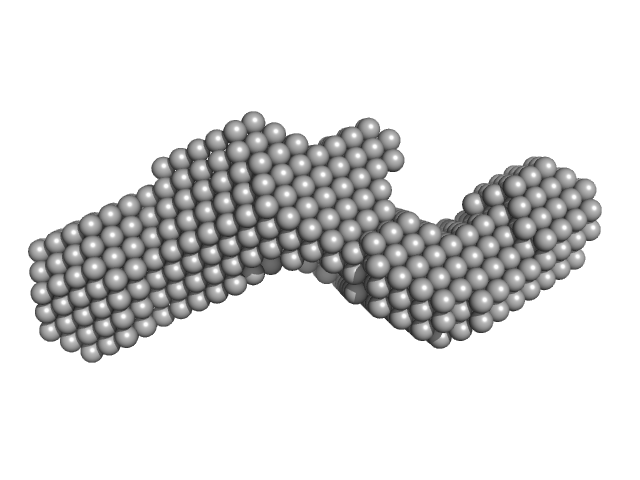
Subdomain SL1 of hepatitis C virus monomer, 15 kDa Hepatitis C virus RNA
|
| Buffer: |
10mM Tris 0.1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 11
|
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.
RNA 23(9):1465-1476 (2017)
Cantero-Camacho Á, Fan L, Wang YX, Gallego J
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.1 |
nm |
|
|
|
|
|
|
|
| Sample: |
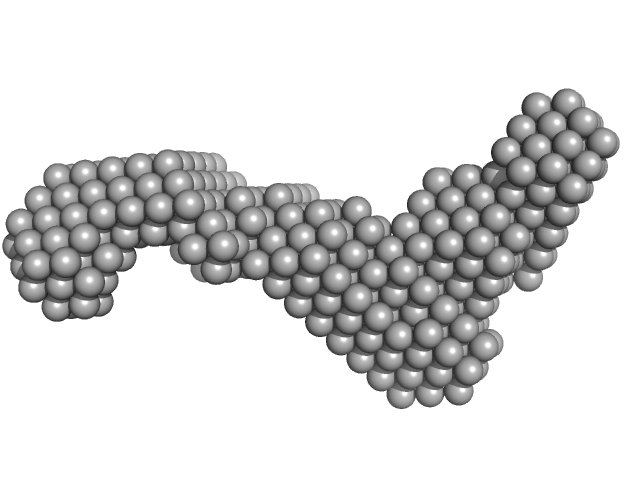
Subdomain SL2' of hepatitis C virus domain 3'X monomer, 18 kDa Hepatitis C virus RNA
|
| Buffer: |
10mM Tris 0.1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 11
|
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.
RNA 23(9):1465-1476 (2017)
Cantero-Camacho Á, Fan L, Wang YX, Gallego J
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.0 |
nm |
|
|
|
|
|
|
|
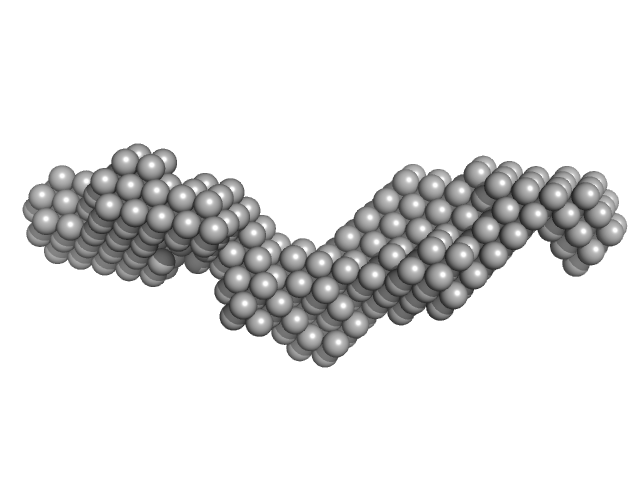
| Sample: |
Domain 3'X of hepatitis C virus monomer, 32 kDa Hepatitis C virus RNA
|
| Buffer: |
10mM Tris 0.1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 11
|
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.
RNA 23(9):1465-1476 (2017)
Cantero-Camacho Á, Fan L, Wang YX, Gallego J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.8 |
nm |
|
|