|
|
|
|
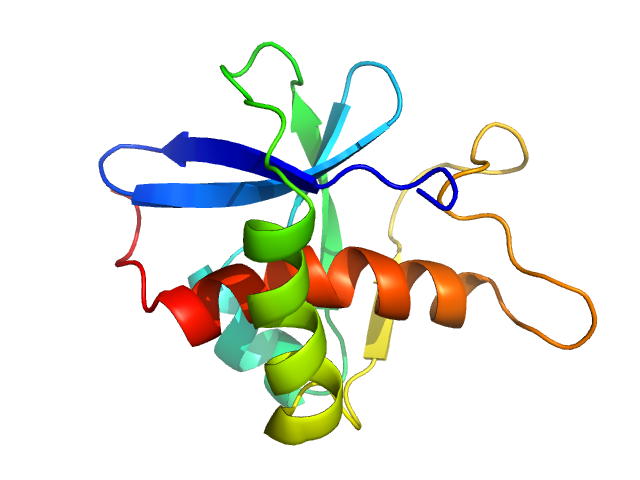
|
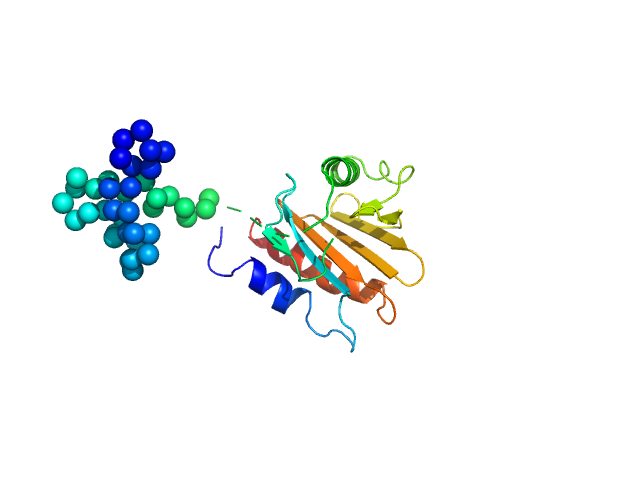
| Sample: |
Genome polyprotein monomer, 13 kDa Turkey avisivirus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TECP, 1% Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 May 2
|
Solution structure and functional characterization of an avian picornavirus 2AH/NC protein
Anna Wehlin
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
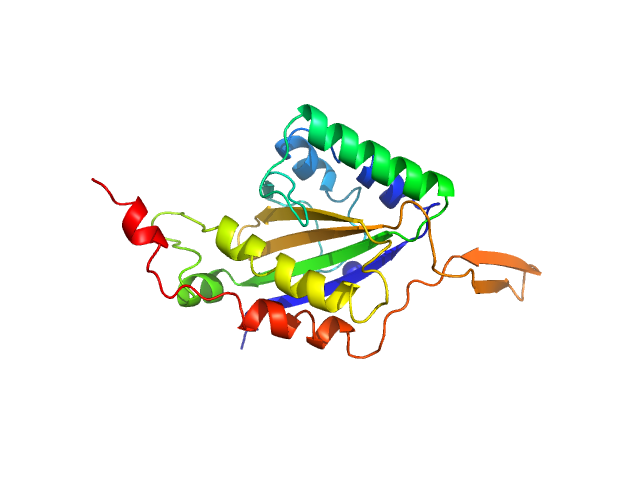
|
| Sample: |
Relaxase monomer, 29 kDa Bacillus subtilis subsp. … protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
Structure of relaxase from Bacillus subtilis conjugative plasmid pLS20 reveals a new mechanism of DNA processing mediated by dimerization
Isidro Crespo
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
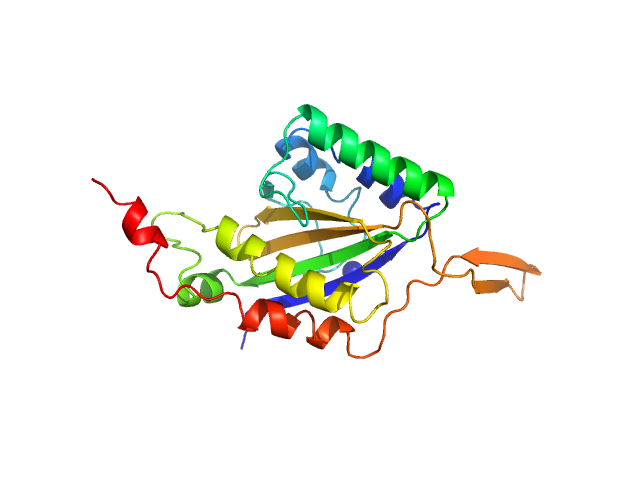
|
| Sample: |
Relaxase dimer, 57 kDa Bacillus subtilis subsp. … protein
Nic sequence, single stranded dimer, 12 kDa Bacillus subtilis DNA
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
Structure of relaxase from Bacillus subtilis conjugative plasmid pLS20 reveals a new mechanism of DNA processing mediated by dimerization
Isidro Crespo
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
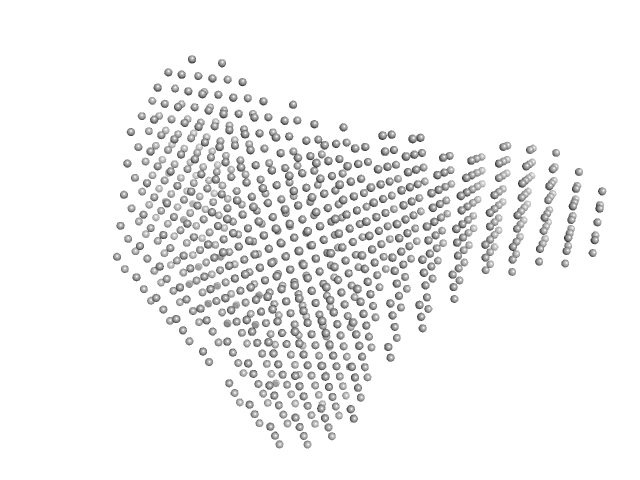
|
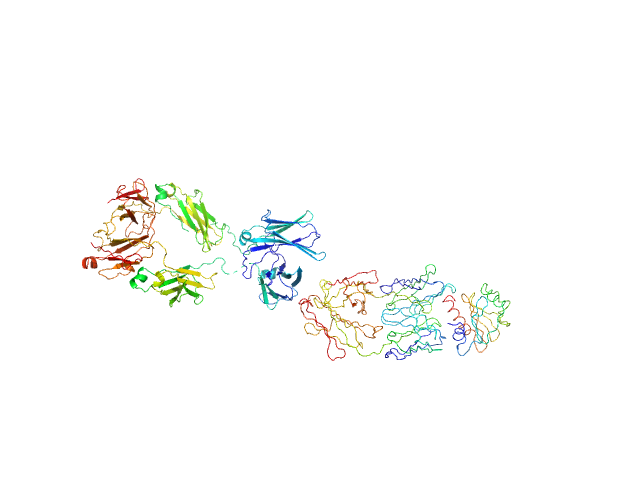
| Sample: |
Murine Immunoglobulin E (IgE) antibodies monomer, 165 kDa Mus musculus protein
Profilin-2 monomer, 18 kDa Hevea brasiliensis protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Oct 3
|
Allergen-induced structural rearrangements in IgE: insights from SAXS and molecular dynamics.
Int J Biol Macromol :147658 (2025)
Gómez-Velasco H, García-Ramírez B, Siliqi D, Graewert MA, Quintero-Martinez A, Ortega E, Rodríguez-Romero A
|
| RgGuinier |
6.0 |
nm |
| Dmax |
19.9 |
nm |
| VolumePorod |
504 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Profilin-2 monomer, 18 kDa Hevea brasiliensis protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Oct 3
|
Allergen-induced structural rearrangements in IgE: insights from SAXS and molecular dynamics.
Int J Biol Macromol :147658 (2025)
Gómez-Velasco H, García-Ramírez B, Siliqi D, Graewert MA, Quintero-Martinez A, Ortega E, Rodríguez-Romero A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Murine Immunoglobulin E (IgE) antibodies monomer, 165 kDa Mus musculus protein
Profilin-2 monomer, 18 kDa Hevea brasiliensis protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2025 Apr 11
|
Allergen-induced structural rearrangements in IgE: insights from SAXS and molecular dynamics.
Int J Biol Macromol :147658 (2025)
Gómez-Velasco H, García-Ramírez B, Siliqi D, Graewert MA, Quintero-Martinez A, Ortega E, Rodríguez-Romero A
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.9 |
nm |
| VolumePorod |
450 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Profilin-2 monomer, 18 kDa Hevea brasiliensis protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2025 Apr 11
|
Allergen-induced structural rearrangements in IgE: insights from SAXS and molecular dynamics.
Int J Biol Macromol :147658 (2025)
Gómez-Velasco H, García-Ramírez B, Siliqi D, Graewert MA, Quintero-Martinez A, Ortega E, Rodríguez-Romero A
|
| RgGuinier |
1.7 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
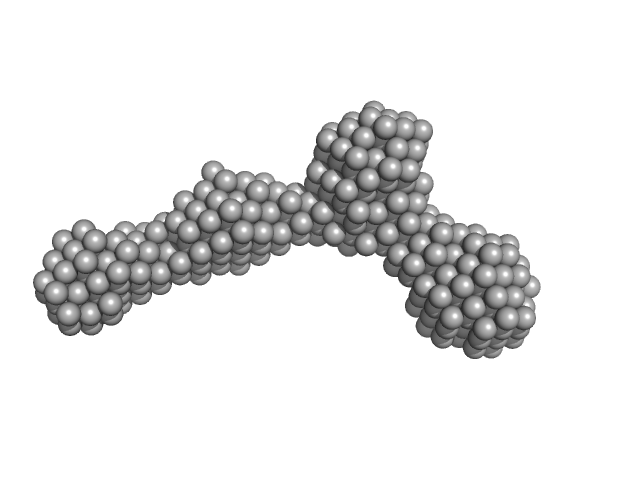
| Sample: |
Murine Immunoglobulin E (IgE) antibodies monomer, 165 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2025 Apr 11
|
Allergen-induced structural rearrangements in IgE: insights from SAXS and molecular dynamics.
Int J Biol Macromol :147658 (2025)
Gómez-Velasco H, García-Ramírez B, Siliqi D, Graewert MA, Quintero-Martinez A, Ortega E, Rodríguez-Romero A
|
| RgGuinier |
5.2 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
420 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
UvrABC system protein B dimer, 156 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH-8, 200 mM NaCl, 2% glycerol, pH: |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2023 Jun 6
|
Inhibition of Mycobacterium tuberculosis UvrB by small molecules: Potent NER disruption and structural insights into dimer conformation.
Int J Biol Macromol :147338 (2025)
Jahan F, Anand S, Kumar S, Pandey G, Siddiqi MI, Krishnan MY, Ramachandran R
|
| RgGuinier |
6.7 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
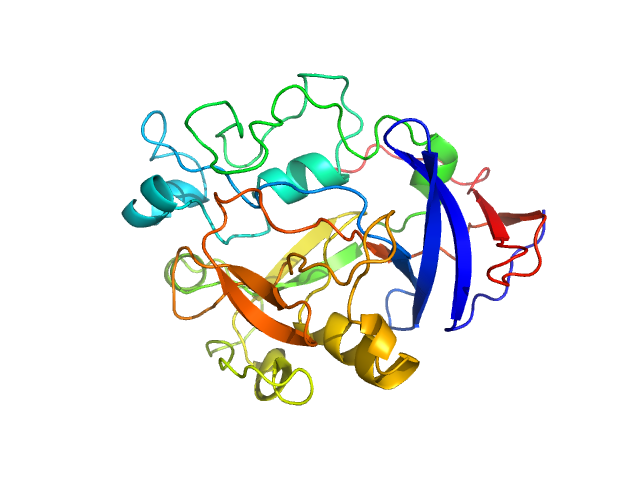
|
| Sample: |
IgA protease monomer, 32 kDa Thomasclavelia ramosa protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Jun 26
|
Structure of the Thomasclavelia ramosa immunoglobulin A protease reveals a modular and minimizable architecture distinct from other immunoglobulin A proteases.
Proc Natl Acad Sci U S A 122(35):e2503549122 (2025)
Tran N, Frenette A, Holyoak T
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|