|
|
|
|
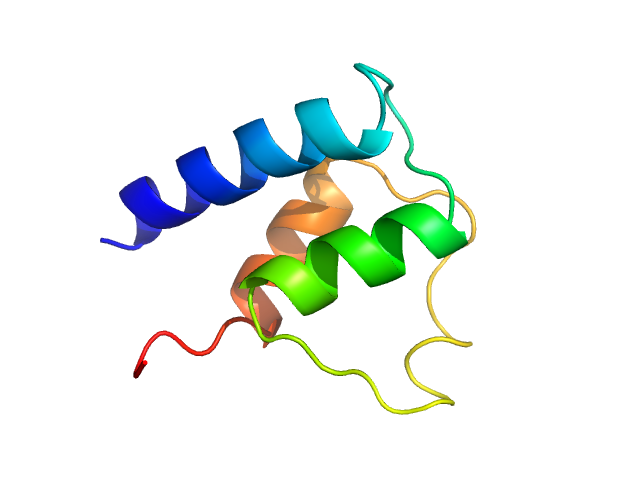
|
| Sample: |
Myosin essential light chain monomer, 9 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 25
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
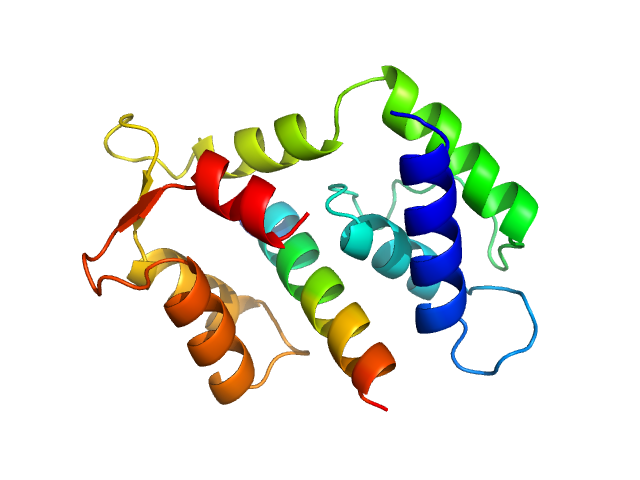
|
| Sample: |
Myosin essential light chain 2 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 3 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 8
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
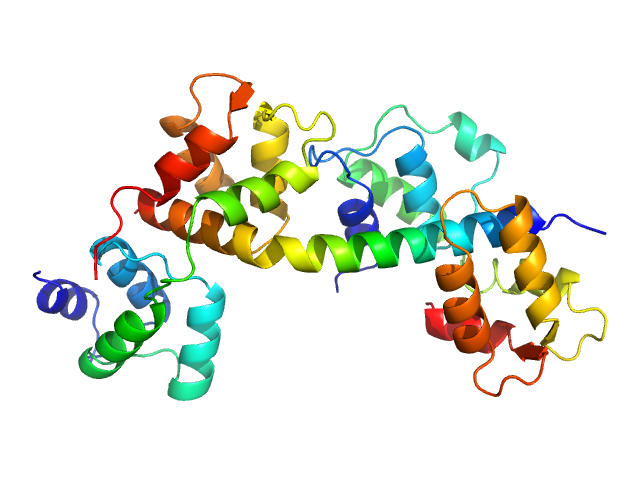
|
| Sample: |
Toxoplasma gondii essential light chain 1 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
Toxoplasma gondii myosin light chain, full length monomer, 24 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
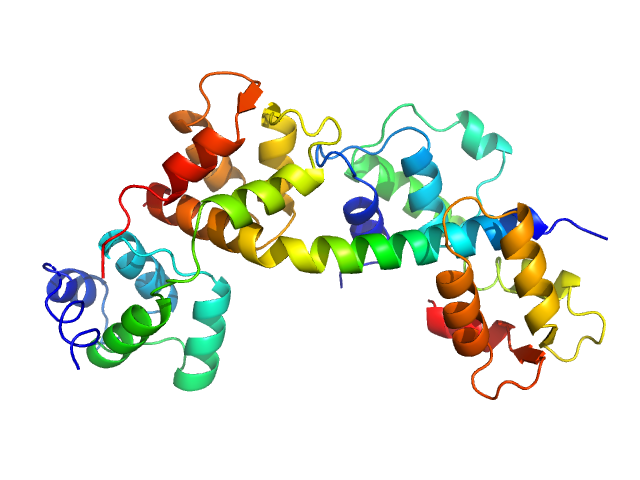
|
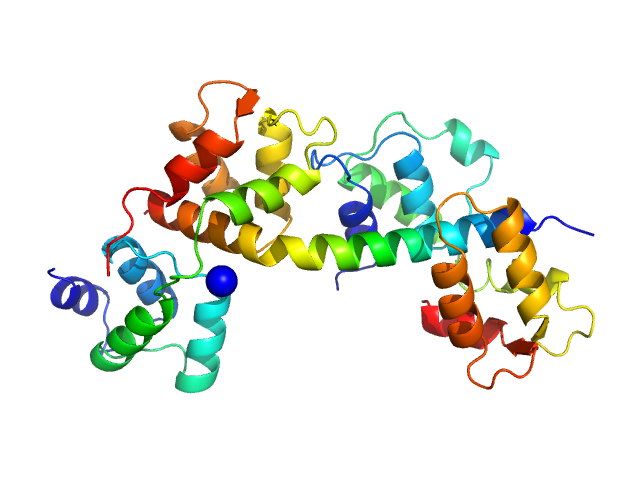
| Sample: |
Myosin light chain TgMLC1, residues 66-210 monomer, 17 kDa Toxoplasma gondii protein
Toxoplasma gondii essential light chain 1 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
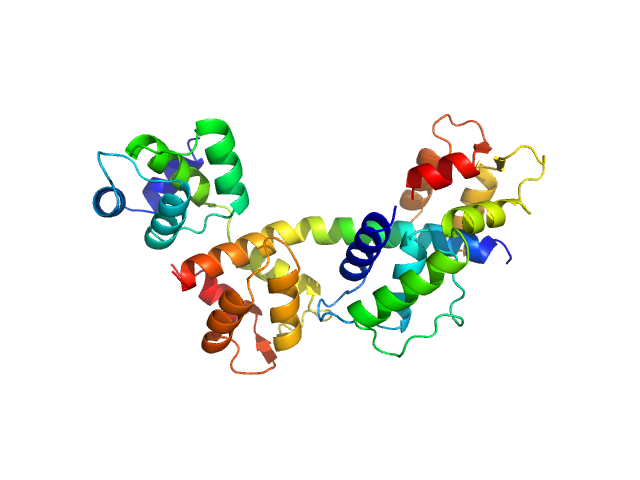
| Sample: |
Toxoplasma gondii essential light chain 1 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
Toxoplasma gondii myosin light chain, residues 70-210 monomer, 17 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
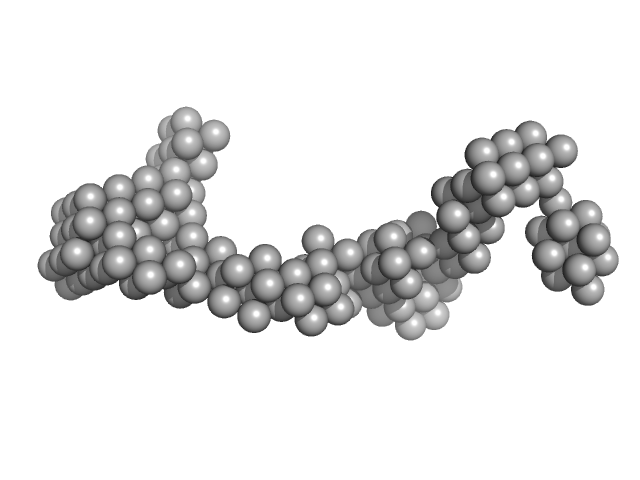
| Sample: |
Myosin essential light chain 2 monomer, 15 kDa Toxoplasma gondii protein
Myosin light chain TgMLC1, residues 66-210 monomer, 17 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillin-1 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID02, ESRF on 2006 Feb 11
|
Transglutaminase-Mediated Cross-Linking of Tropoelastin to Fibrillin Stabilises the Elastin Precursor Prior to Elastic Fibre Assembly.
J Mol Biol 432(21):5736-5751 (2020)
Lockhart-Cairns MP, Newandee H, Thomson J, Weiss AS, Baldock C, Tarakanova A
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Mce-family protein Mce1A monomer monomer, 17 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris, 350 mM NaCl, 10% Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 May 10
|
Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis
IUCrJ 8(5) (2021)
Asthana P, Singh D, Pedersen J, Hynönen M, Sulu R, Murthy A, Laitaoja M, Jänis J, Riley L, Venkatesan R
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Mce-family protein Mce4A monomer, 15 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50mM MOPS, 350mM NaCl, 10% Glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 May 10
|
Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis
IUCrJ 8(5) (2021)
Asthana P, Singh D, Pedersen J, Hynönen M, Sulu R, Murthy A, Laitaoja M, Jänis J, Riley L, Venkatesan R
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
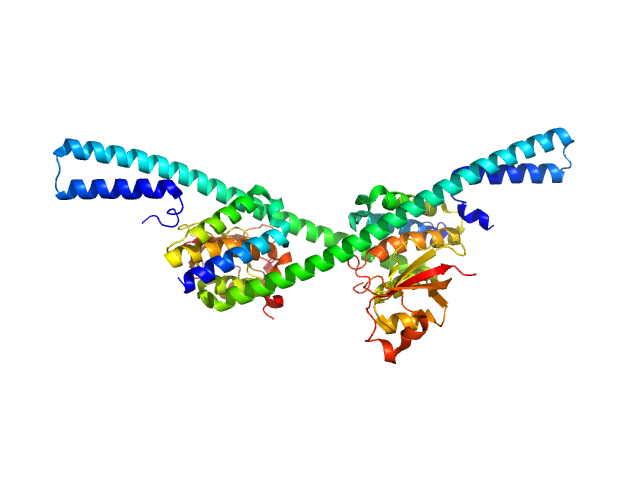
| Sample: |
Replicase polyprotein 1a - nsp7, 18 kDa Severe acute respiratory … protein
Replicase polyprotein 1a - nsp8, 44 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, pH 8.0, 100 mM NaCl, 4 mM DTT, 4 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 19
|
Hallmarks of Alpha-
and Betacoronavirus
non-structural protein 7+8 complexes
Science Advances 7(10):eabf1004 (2021)
Krichel B, Bylapudi G, Schmidt C, Blanchet C, Schubert R, Brings L, Koehler M, Zenobi R, Svergun D, Lorenzen K, Madhugiri R, Ziebuhr J, Uetrecht C
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
92 |
nm3 |
|
|