|
|
|
|
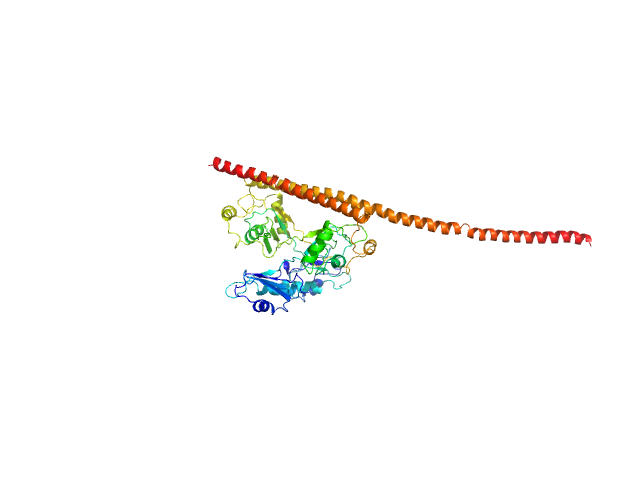
|
| Sample: |
Splicing factor, proline- and glutamine-rich monomer, 38 kDa Homo sapiens protein
Non-POU domain-containing octamer-binding protein monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 8
|
Structural basis of the zinc-induced cytoplasmic aggregation of the RNA-binding protein SFPQ
Nucleic Acids Research 48(6):3356-3365 (2020)
Huang J, Ringuet M, Whitten A, Caria S, Lim Y, Badhan R, Anggono V, Lee M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
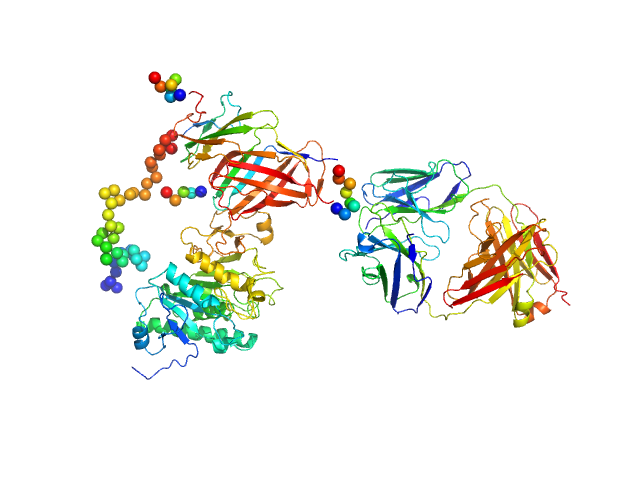
|
| Sample: |
Lipoprotein lipase monomer, 50 kDa Homo sapiens protein
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 monomer, 15 kDa Homo sapiens protein
Monoclonal Antibody Fragment 5D2 monomer, 47 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris, 150 mM NaCl, 4 mM CaCL2, 10% (v/v) Glycerol, 0.05% 0.8mM CHAPS, ,0.05 % (v/v) NaN3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 14
|
Unfolding of monomeric lipoprotein lipase by ANGPTL4: Insight into the regulation of plasma triglyceride metabolism
Proceedings of the National Academy of Sciences :201920202 (2020)
Kristensen K, Leth-Espensen K, Mertens H, Birrane G, Meiyappan M, Olivecrona G, Jørgensen T, Young S, Ploug M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
187 |
nm3 |
|
|
|
|
|
|
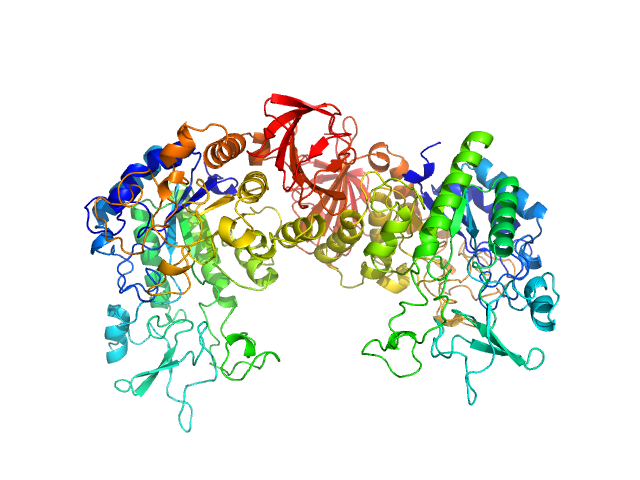
|
| Sample: |
Binary toxin receptor Cqm1 protein dimer, 129 kDa synthetic construct protein
|
| Buffer: |
25 mM HEPES, pH 7.5, 25 mM NaCl, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-I facility, Dhruva Reactor, Bhabha Atomic Research Centre on 2019 Apr 24
|
Small-angle neutron scattering studies suggest the mechanism of BinAB protein internalization
IUCrJ 7(2) (2020)
Sharma M, Aswal V, Kumar V, Chidambaram R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.7 |
nm |
|
|
|
|
|
|
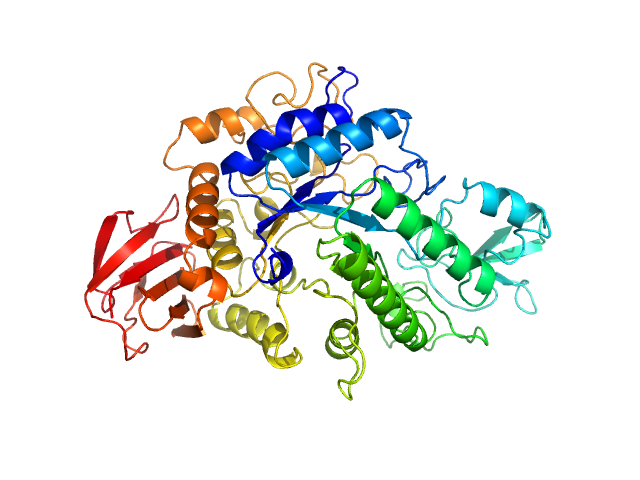
|
| Sample: |
Binary toxin receptor Cqm1 protein monomer, 65 kDa synthetic construct protein
Deuterated BinB component of mosquito-larvicidal Binary toxin monomer, 56 kDa synthetic construct protein
|
| Buffer: |
25 mM HEPES, pH 7.5, 25 mM NaCl, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-I facility, Dhruva Reactor, Bhabha Atomic Research Centre on 2019 Apr 23
|
Small-angle neutron scattering studies suggest the mechanism of BinAB protein internalization
IUCrJ 7(2) (2020)
Sharma M, Aswal V, Kumar V, Chidambaram R
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.3 |
nm |
|
|
|
|
|
|
|
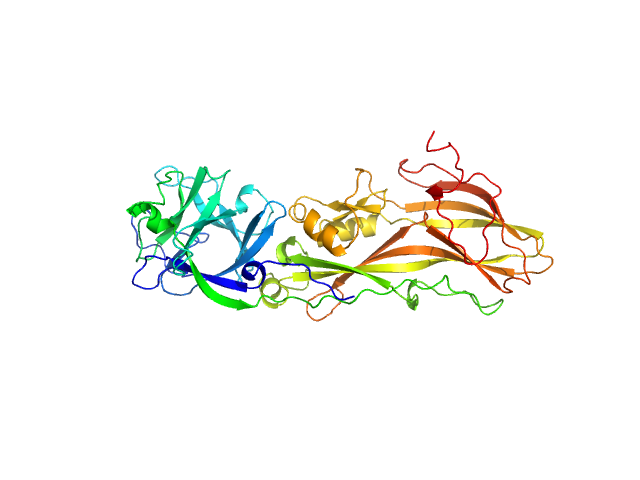
| Sample: |
Lysinibacillus Mosquito-larvicidal receptor binding component monomer, 53 kDa synthetic construct protein
|
| Buffer: |
25 mM HEPES, pH 7.5, 25 mM NaCl, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-I facility, Dhruva Reactor, Bhabha Atomic Research Centre on 2019 Apr 24
|
Small-angle neutron scattering studies suggest the mechanism of BinAB protein internalization
IUCrJ 7(2) (2020)
Sharma M, Aswal V, Kumar V, Chidambaram R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.9 |
nm |
|
|
|
|
|
|
|
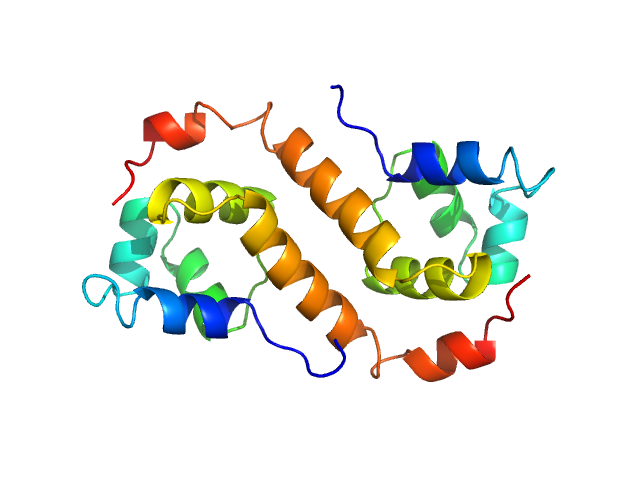
| Sample: |
Uncharacterized protein dimer, 22 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 5% (v/v) glycerol, and 1 mM PMSF, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Dec 21
|
Structural Insights Into the Transcriptional Regulation of HigBA Toxin–Antitoxin System by Antitoxin HigA in Pseudomonas aeruginosa
Frontiers in Microbiology 10 (2020)
Liu Y, Gao Z, Liu G, Geng Z, Dong Y, Zhang H
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
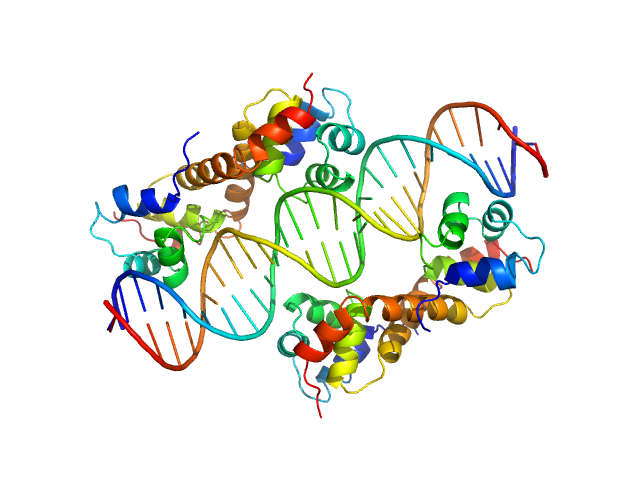
| Sample: |
Uncharacterized protein dimer, 22 kDa Pseudomonas aeruginosa protein
DNA Duplex dimer, 20 kDa DNA
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 5% (v/v) glycerol, and 1 mM PMSF, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Sep 19
|
Structural Insights Into the Transcriptional Regulation of HigBA Toxin–Antitoxin System by Antitoxin HigA in Pseudomonas aeruginosa
Frontiers in Microbiology 10 (2020)
Liu Y, Gao Z, Liu G, Geng Z, Dong Y, Zhang H
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
|
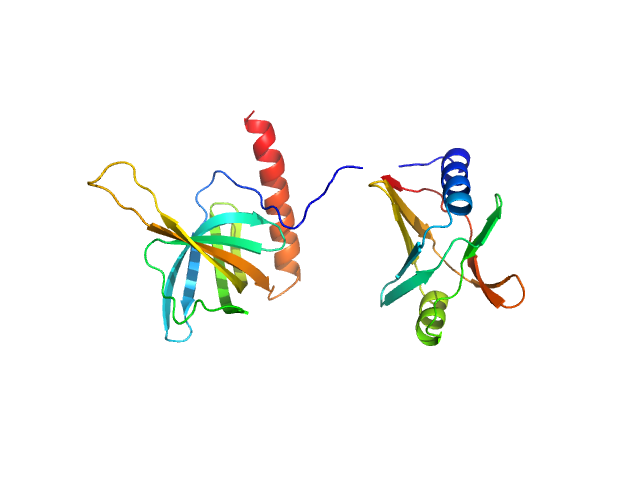
| Sample: |
Flagellar brake protein YcgR monomer, 29 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES, 150mM NaCl, 10% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Jan 4
|
Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility in Escherichia coli.
J Biol Chem 295(3):808-821 (2020)
Hou YJ, Yang WS, Hong Y, Zhang Y, Wang DC, Li DF
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Flagellar brake protein YcgR in complex with c-di-GMP monomer, 29 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES, 150mM NaCl, 10% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Jan 4
|
Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility in Escherichia coli.
J Biol Chem 295(3):808-821 (2020)
Hou YJ, Yang WS, Hong Y, Zhang Y, Wang DC, Li DF
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
De novo protein WA20 dimer, 25 kDa de novo protein protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 200 mM ArgHCl, 10% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-6A, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2016 Feb 26
|
Hyperstable De Novo Protein with a Dimeric Bisecting Topology.
ACS Synth Biol (2020)
Kimura N, Mochizuki K, Umezawa K, Hecht MH, Arai R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
|
|