|
|
|
|
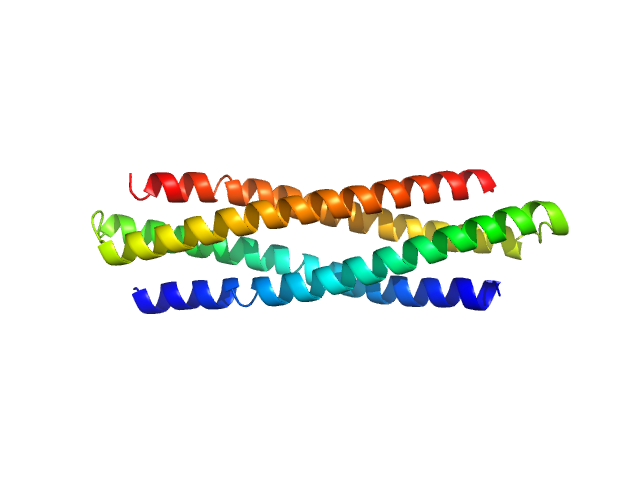
|
| Sample: |
Hyperstable de novo protein Super WA20 dimer, 25 kDa de novo protein protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 200 mM ArgHCl, 10% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-6A, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2016 Feb 26
|
Hyperstable De Novo Protein with a Dimeric Bisecting Topology.
ACS Synth Biol (2020)
Kimura N, Mochizuki K, Umezawa K, Hecht MH, Arai R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDGS5_fit1_model1.png)
|
| Sample: |
MvaT(mutant) dimer, 28 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
20 mM Bis-Tris 50 mM KCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 11
|
Structural basis for osmotic regulation of the DNA binding properties of H-NS proteins.
Nucleic Acids Res (2020)
Qin L, Bdira FB, Sterckx YGJ, Volkov AN, Vreede J, Giachin G, van Schaik P, Ubbink M, Dame RT
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDGT5_fit1_model1.png)
|
| Sample: |
MvaT(mutant) dimer, 28 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
20 mM Bis-Tris 300 mM KCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 11
|
Structural basis for osmotic regulation of the DNA binding properties of H-NS proteins.
Nucleic Acids Res (2020)
Qin L, Bdira FB, Sterckx YGJ, Volkov AN, Vreede J, Giachin G, van Schaik P, Ubbink M, Dame RT
|
| RgGuinier |
3.8 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
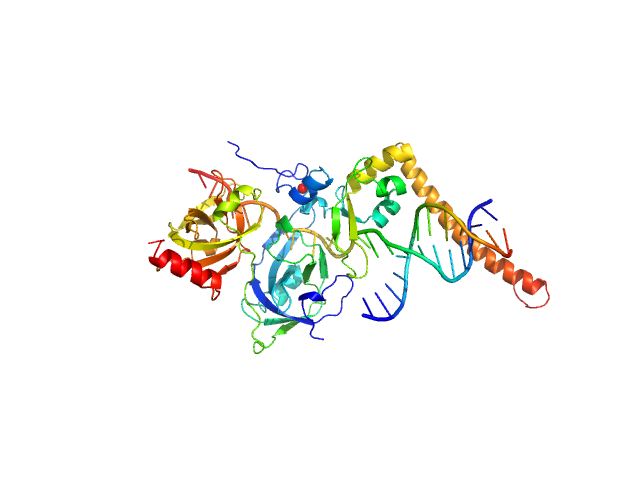
|
| Sample: |
DNA repair protein complementing XP-A cells monomer, 17 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 27 kDa Homo sapiens protein
3-prime Nucleotide Excision Repair Junction Model Substrate monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 17 Nov 2
|
A key interaction with RPA orients XPA in NER complexes.
Nucleic Acids Res (2020)
Topolska-Woś AM, Sugitani N, Cordoba JJ, Le Meur KV, Le Meur RA, Kim HS, Yeo JE, Rosenberg D, Hammel M, Schärer OD, Chazin WJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
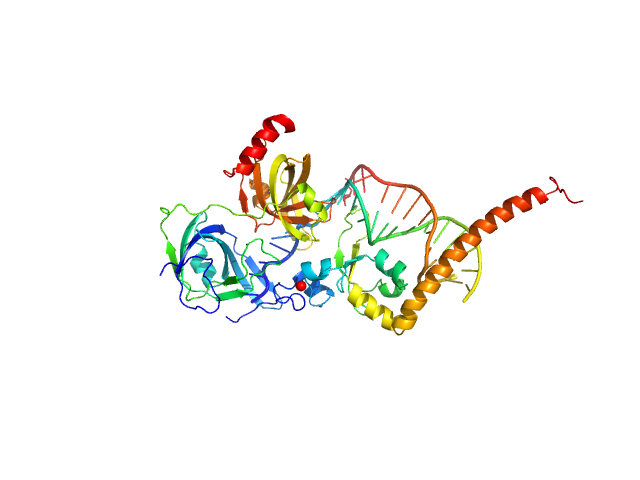
|
| Sample: |
DNA repair protein complementing XP-A cells monomer, 17 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 27 kDa Homo sapiens protein
5-prime Nucleotide Excision Repair Junction Model Substrate monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 4
|
A key interaction with RPA orients XPA in NER complexes.
Nucleic Acids Res (2020)
Topolska-Woś AM, Sugitani N, Cordoba JJ, Le Meur KV, Le Meur RA, Kim HS, Yeo JE, Rosenberg D, Hammel M, Schärer OD, Chazin WJ
|
| RgGuinier |
2.9 |
nm |
| Dmax |
97.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
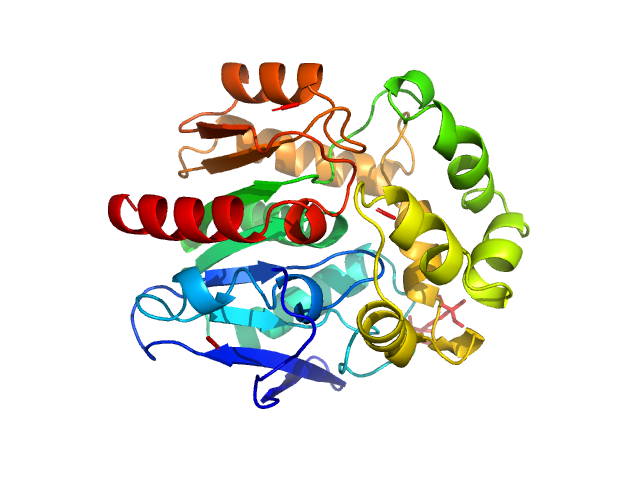
|
| Sample: |
Haloalkane dehalogenase variant DhaA115 -monomeric fraction monomer, 34 kDa Rhodococcus rhodochrous protein
|
| Buffer: |
50 mM potassium phosphate buffer (41 mM K₂HPO₄, 9mM KH₂PO₄), pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2019 Aug 22
|
Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst
Chemical Science 11(41):11162-11178 (2020)
Markova K, Chmelova K, Marques S, Carpentier P, Bednar D, Damborsky J, Marek M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
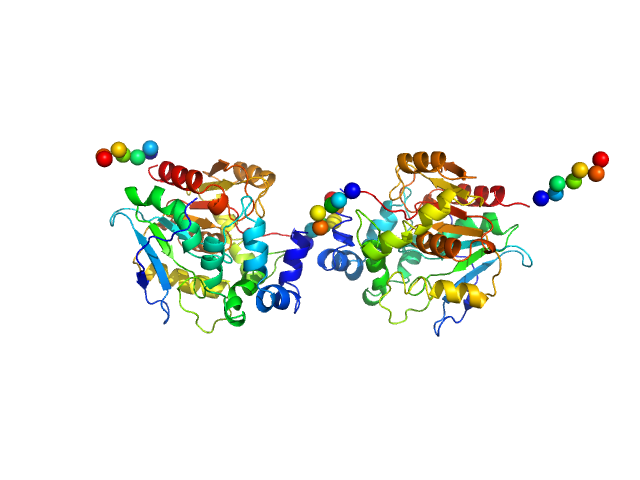
| Sample: |
Haloalkane dehalogenase variant DhaA115 - dimeric fraction dimer, 69 kDa Rhodococcus rhodochrous protein
|
| Buffer: |
50 mM potassium phosphate buffer (41 mM K₂HPO₄, 9mM KH₂PO₄), pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2019 Aug 22
|
Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst
Chemical Science 11(41):11162-11178 (2020)
Markova K, Chmelova K, Marques S, Carpentier P, Bednar D, Damborsky J, Marek M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
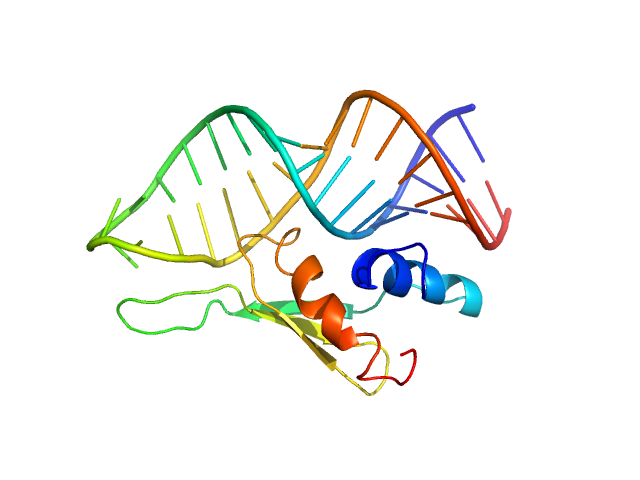
| Sample: |
Double-stranded RNA-binding protein Staufen homolog 1 - RNA binding domain 4 monomer, 8 kDa Homo sapiens protein
ADP-ribosylation factor1 - short monomer, 11 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 3.5 mM 2-mercaptoethanol, pH: 6.8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2015 Dec 10
|
Staufen1 reads out structure and sequence features in ARF1 dsRNA for target recognition.
Nucleic Acids Res (2019)
Yadav DK, Zigáčková D, Zlobina M, Klumpler T, Beaumont C, Kubíčková M, Vaňáčová Š, Lukavsky PJ
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
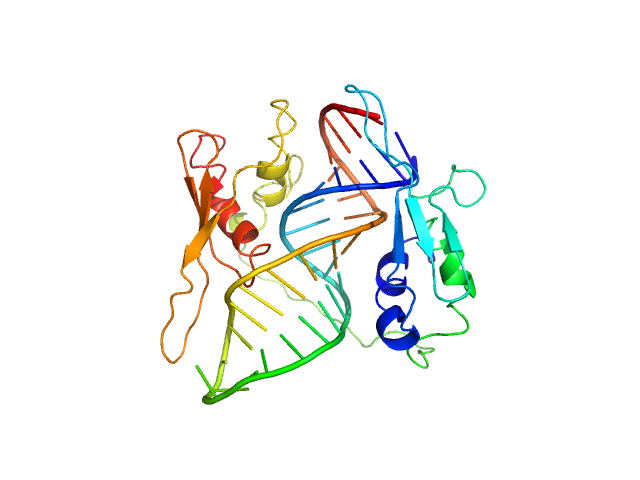
| Sample: |
ADP-ribosylation factor1 - short monomer, 11 kDa Homo sapiens RNA
Double-stranded RNA-binding protein Staufen homolog 1 - RNA binding domain 3 and 4 monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate buffer, 300 mM NaCl, 500 mM imidazole, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2018 May 29
|
Staufen1 reads out structure and sequence features in ARF1 dsRNA for target recognition.
Nucleic Acids Res (2019)
Yadav DK, Zigáčková D, Zlobina M, Klumpler T, Beaumont C, Kubíčková M, Vaňáčová Š, Lukavsky PJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
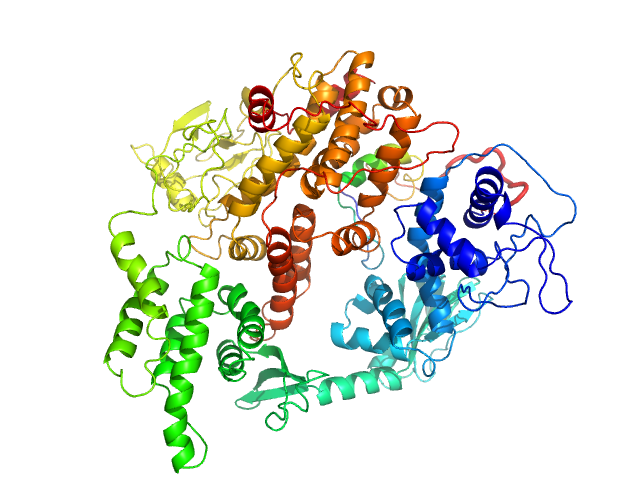
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 100 kDa Homo sapiens protein
|
| Buffer: |
1mM EDTA, 10mM DTT, 500mM NaCl, and 10mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2012 Sep 7
|
Conformational States of Exchange Protein Directly Activated by cAMP (EPAC1) Revealed by Ensemble Modeling and Integrative Structural Biology.
Cells 9(1) (2019)
White MA, Tsalkova T, Mei FC, Cheng X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|