|
|
|
|

|
| Sample: |
Zinc finger and BTB domain-containing protein 38 monomer, 17 kDa Homo sapiens protein
Methylated C-terminal ZBTB38 binding sequence monomer, 17 kDa DNA
|
| Buffer: |
10 mM Tris, 1 mM tris(2-carboxy-ethyl)phosphine (TCEP), 0.05% NaN3, 10% D2O, pH: 6.8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, Department of Chemistry, University of Utah on 2016 Nov 11
|
The C-Terminal Zinc Fingers of ZBTB38 are Novel Selective Readers of DNA Methylation.
J Mol Biol 430(3):258-271 (2018)
Pozner A, Hudson NO, Trewhella J, Terooatea TW, Miller SA, Buck-Koehntop BA
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
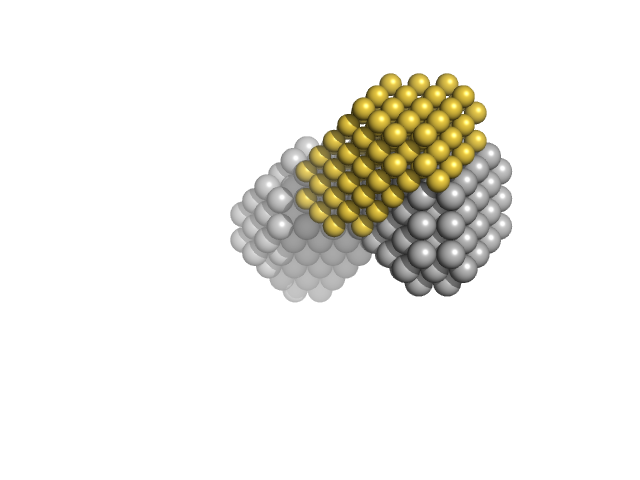
|
| Sample: |
Basic domain of telomeric repeat-binding factor 2 monomer, 5 kDa Homo sapiens protein
Telomere DNA duplex monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 50 mM LiCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 May 3
|
Basic domain of telomere guardian TRF2 reduces D-loop unwinding whereas Rap1 restores it.
Nucleic Acids Res 45(21):12170-12180 (2017)
Necasová I, Janoušková E, Klumpler T, Hofr C
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
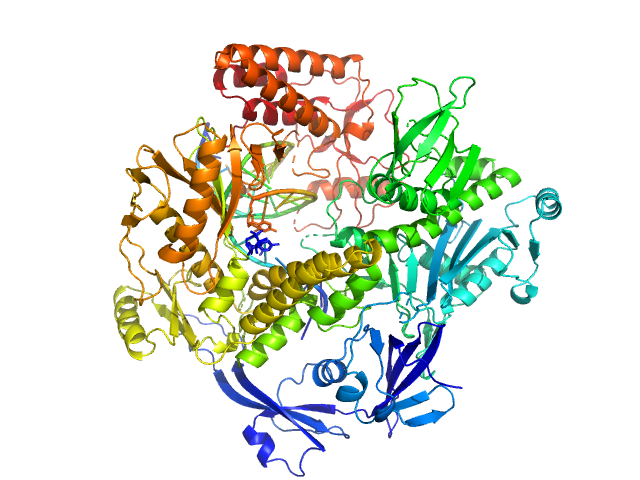
| Sample: |
DNA polymerase E9 exo- mutant monomer, 117 kDa Vaccinia virus protein
DNA oligomer template-primer hairpin monomer, 9 kDa synthetic construct DNA
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 4 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 12
|
The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun 8(1):1455 (2017)
Tarbouriech N, Ducournau C, Hutin S, Mas PJ, Man P, Forest E, Hart DJ, Peyrefitte CN, Burmeister WP, Iseni F
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
40bp long dsDNA-Sa Oligonucleotide monomer, 25 kDa DNA
Wild-type archaeal biofilm regulator 1 (ABfR1: Transcriptional regulator ArsR family). dimer, 26 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
0.5 x Tris/Borate/EDTA (TBE), pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 27
|
Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius.
Mol Microbiol 105(5):777-793 (2017)
Li L, Banerjee A, Bischof LF, Maklad HR, Hoffmann L, Henche AL, Veliz F, Bildl W, Schulte U, Orell A, Essen LO, Peeters E, Albers SV
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 30 kDa Homo sapiens protein
Poly U 15mer monomer, 5 kDa RNA
|
| Buffer: |
10 mM Potassium Phosphate 50 mM NaCl 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2016 Apr 28
|
Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew Chem Int Ed Engl 56(32):9322-9325 (2017)
Sonntag M, Jagtap PKA, Simon B, Appavou MS, Geerlof A, Stehle R, Gabel F, Hennig J, Sattler M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly U 15mer monomer, 5 kDa RNA
Nucleolysin TIA-1 isoform p40 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
10 mM Potassium Phosphate 50 mM NaCl 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2015 Dec 1
|
Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew Chem Int Ed Engl 56(32):9322-9325 (2017)
Sonntag M, Jagtap PKA, Simon B, Appavou MS, Geerlof A, Stehle R, Gabel F, Hennig J, Sattler M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly U 15mer monomer, 5 kDa RNA
Nucleolysin TIA-1 isoform p40 monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
10 mM Potassium Phosphate 50 mM NaCl 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2016 May 4
|
Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew Chem Int Ed Engl 56(32):9322-9325 (2017)
Sonntag M, Jagtap PKA, Simon B, Appavou MS, Geerlof A, Stehle R, Gabel F, Hennig J, Sattler M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
|
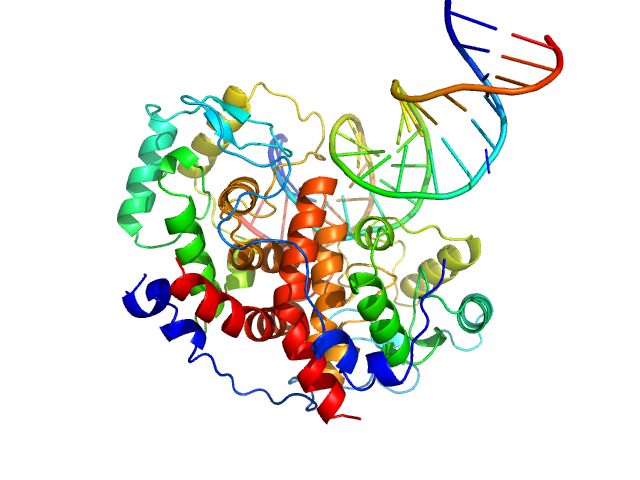
| Sample: |
Major viral transcription factor ICP4 dimer, 49 kDa Human alphaherpesvirus 1 protein
Immediate Early 3 dimer, 12 kDa DNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Jul 27
|
The herpes viral transcription factor ICP4 forms a novel DNA recognition complex.
Nucleic Acids Res 45(13):8064-8078 (2017)
Tunnicliffe RB, Lockhart-Cairns MP, Levy C, Mould AP, Jowitt TA, Sito H, Baldock C, Sandri-Goldin RM, Golovanov AP
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
RNA (ACUCCUUUUU) monomer, 1 kDa RNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 May 27
|
TIA-1 RRM23 binding and recognition of target oligonucleotides.
Nucleic Acids Res 45(8):4944-4957 (2017)
Waris S, García-Mauriño SM, Sivakumaran A, Beckham SA, Loughlin FE, Gorospe M, Díaz-Moreno I, Wilce MCJ, Wilce JA
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
DNA (ACTCCTTTTT) monomer, 1 kDa DNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 May 27
|
TIA-1 RRM23 binding and recognition of target oligonucleotides.
Nucleic Acids Res 45(8):4944-4957 (2017)
Waris S, García-Mauriño SM, Sivakumaran A, Beckham SA, Loughlin FE, Gorospe M, Díaz-Moreno I, Wilce MCJ, Wilce JA
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
32 |
nm3 |
|
|