|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDGS5_fit1_model1.png)
|
| Sample: |
MvaT(mutant) dimer, 28 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
20 mM Bis-Tris 50 mM KCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 11
|
Structural basis for osmotic regulation of the DNA binding properties of H-NS proteins.
Nucleic Acids Res (2020)
Qin L, Bdira FB, Sterckx YGJ, Volkov AN, Vreede J, Giachin G, van Schaik P, Ubbink M, Dame RT
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDGT5_fit1_model1.png)
|
| Sample: |
MvaT(mutant) dimer, 28 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
20 mM Bis-Tris 300 mM KCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 11
|
Structural basis for osmotic regulation of the DNA binding properties of H-NS proteins.
Nucleic Acids Res (2020)
Qin L, Bdira FB, Sterckx YGJ, Volkov AN, Vreede J, Giachin G, van Schaik P, Ubbink M, Dame RT
|
| RgGuinier |
3.8 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Resistance to inhibitors of cholinesterase 8 homolog A monomer, 56 kDa Rattus norvegicus protein
Guanine nucleotide-binding protein G(i) subunit alpha-1 monomer, 38 kDa Rattus norvegicus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 30
|
Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to Gαi1
Nature Communications 11(1) (2020)
McClelland L, Zhang K, Mou T, Johnston J, Yates-Hansen C, Li S, Thomas C, Doukov T, Triest S, Wohlkonig A, Tall G, Steyaert J, Chiu W, Sprang S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
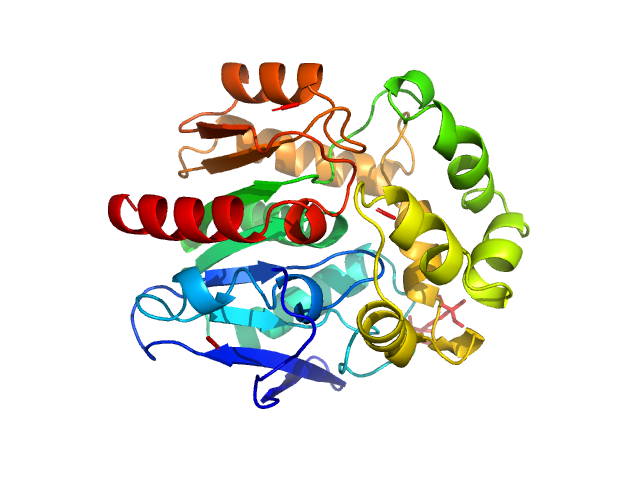
|
| Sample: |
Haloalkane dehalogenase variant DhaA115 -monomeric fraction monomer, 34 kDa Rhodococcus rhodochrous protein
|
| Buffer: |
50 mM potassium phosphate buffer (41 mM K₂HPO₄, 9mM KH₂PO₄), pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2019 Aug 22
|
Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst
Chemical Science 11(41):11162-11178 (2020)
Markova K, Chmelova K, Marques S, Carpentier P, Bednar D, Damborsky J, Marek M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
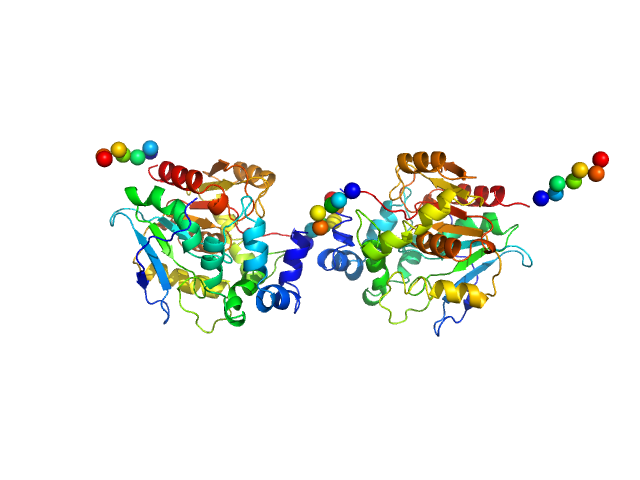
|
| Sample: |
Haloalkane dehalogenase variant DhaA115 - dimeric fraction dimer, 69 kDa Rhodococcus rhodochrous protein
|
| Buffer: |
50 mM potassium phosphate buffer (41 mM K₂HPO₄, 9mM KH₂PO₄), pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2019 Aug 22
|
Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst
Chemical Science 11(41):11162-11178 (2020)
Markova K, Chmelova K, Marques S, Carpentier P, Bednar D, Damborsky J, Marek M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 100 mM NaCl, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Oct 28
|
Polymorphic Protective Dps-DNA Co-Crystals by Cryo Electron Tomography and Small Angle X-Ray Scattering.
Biomolecules 10(1) (2019)
Kamyshinsky R, Chesnokov Y, Dadinova L, Mozhaev A, Orlov I, Petoukhov M, Orekhov A, Shtykova E, Vasiliev A
|
|
|
|
|
|
|
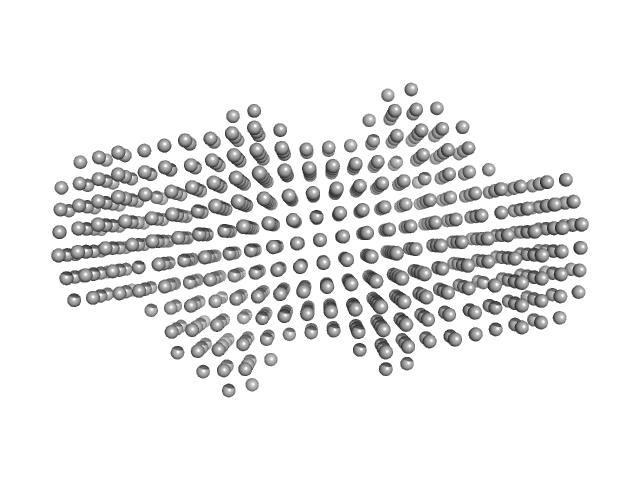
|
| Sample: |
P. maculata perivitellin 2 dimer, 188 kDa Pomacea maculata protein
|
| Buffer: |
20 mM Tris, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2015 Mar 26
|
Exaptation of two ancient immune proteins into a new dimeric pore-forming toxin in snails.
J Struct Biol 211(2):107531 (2020)
Giglio ML, Ituarte S, Milesi V, Dreon MS, Brola TR, Caramelo J, Ip JCH, Maté S, Qiu JW, Otero LH, Heras H
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
267 |
nm3 |
|
|
|
|
|
|
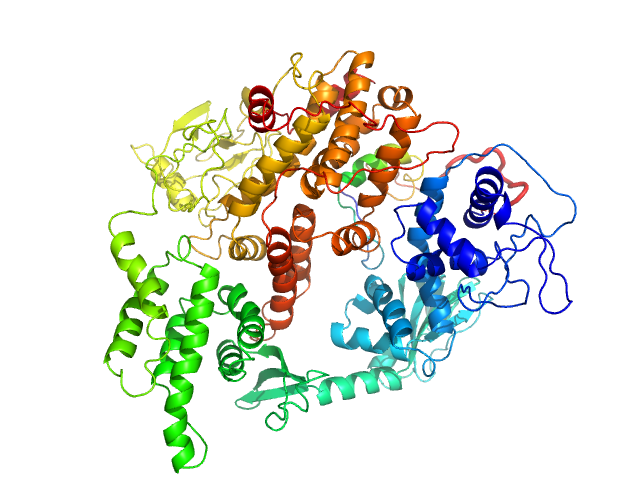
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 100 kDa Homo sapiens protein
|
| Buffer: |
1mM EDTA, 10mM DTT, 500mM NaCl, and 10mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2012 Sep 7
|
Conformational States of Exchange Protein Directly Activated by cAMP (EPAC1) Revealed by Ensemble Modeling and Integrative Structural Biology.
Cells 9(1) (2019)
White MA, Tsalkova T, Mei FC, Cheng X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
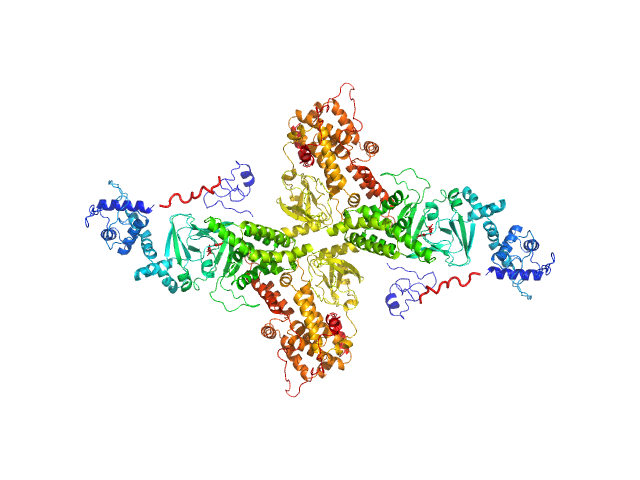
|
| Sample: |
Rap guanine nucleotide exchange factor 3 (dimer) dimer, 200 kDa Homo sapiens protein
|
| Buffer: |
1mM EDTA, 10mM DTT, 500mM NaCl, 1mM cAMP, and 10mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2012 Jan 30
|
Conformational States of Exchange Protein Directly Activated by cAMP (EPAC1) Revealed by Ensemble Modeling and Integrative Structural Biology.
Cells 9(1) (2019)
White MA, Tsalkova T, Mei FC, Cheng X
|
| RgGuinier |
5.3 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
415 |
nm3 |
|
|
|
|
|
|
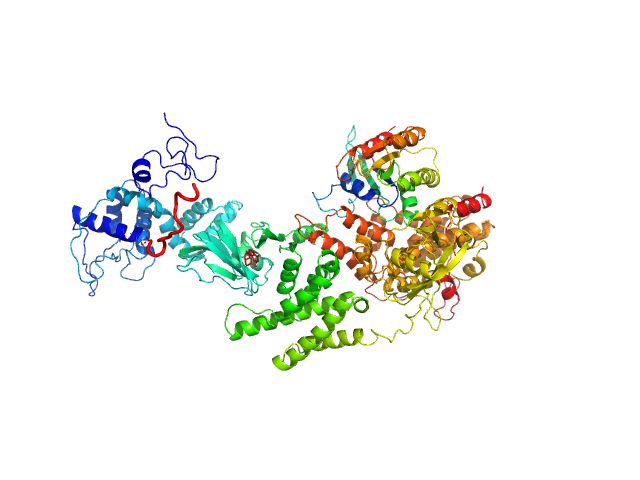
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 100 kDa Homo sapiens protein
RAS related protein 1b monomer, 18 kDa Mus musculus protein
|
| Buffer: |
1mM EDTA, 10mM DTT, 500mM NaCl, 1mM cAMP, and 10mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2013 Apr 1
|
Conformational States of Exchange Protein Directly Activated by cAMP (EPAC1) Revealed by Ensemble Modeling and Integrative Structural Biology.
Cells 9(1) (2019)
White MA, Tsalkova T, Mei FC, Cheng X
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
207 |
nm3 |
|
|