|
|
|
|
|
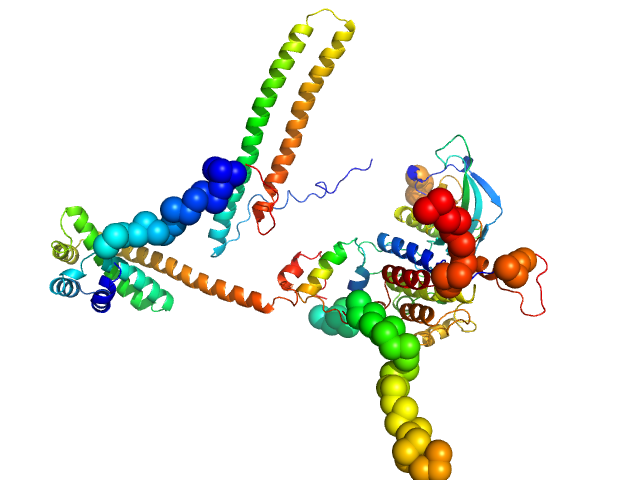
| Sample: |
Hsp90 co-chaperone Cdc37 monomer, 44 kDa Homo sapiens protein
Fibroblast growth factor receptor 3 monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Disease Variants of FGFR3 Reveal Molecular Basis for the Recognition and Additional Roles for Cdc37 in Hsp90 Chaperone System.
Structure 26(3):446-458.e8 (2018)
Bunney TD, Inglis AJ, Sanfelice D, Farrell B, Kerr CJ, Thompson GS, Masson GR, Thiyagarajan N, Svergun DI, Williams RL, Breeze AL, Katan M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fibroblast growth factor receptor 3 monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Disease Variants of FGFR3 Reveal Molecular Basis for the Recognition and Additional Roles for Cdc37 in Hsp90 Chaperone System.
Structure 26(3):446-458.e8 (2018)
Bunney TD, Inglis AJ, Sanfelice D, Farrell B, Kerr CJ, Thompson GS, Masson GR, Thiyagarajan N, Svergun DI, Williams RL, Breeze AL, Katan M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Saposin-a monomer, 9 kDa Homo sapiens protein
1,2-dioleoyl-sn-glycero-3-phosphocholine, 47 kDa synthetic construct
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 15
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
4.1 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Saposin-a monomer, 9 kDa Homo sapiens protein
L-α-phosphatidylinositol (soy), 52 kDa Glycine max
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 15
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
3.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Saposin-a monomer, 9 kDa Homo sapiens protein
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol), 46 kDa synthetic construct
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 15
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
4.1 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Saposin-a monomer, 9 kDa Homo sapiens protein
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine, 47 kDa synthetic construct
Mechanosensitive channel T2 heptamer, 230 kDa Thermoplasma volcanium protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 4
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
5.4 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
686 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Saposin-a monomer, 9 kDa Homo sapiens protein
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine, 47 kDa synthetic construct
Dipeptide and tripeptide permease A monomer, 51 kDa Escherichia coli protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 4
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
288 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Zinc finger and BTB domain-containing protein 38 monomer, 17 kDa Homo sapiens protein
Methylated C-terminal ZBTB38 binding sequence monomer, 17 kDa DNA
|
| Buffer: |
10 mM Tris, 1 mM tris(2-carboxy-ethyl)phosphine (TCEP), 0.05% NaN3, 10% D2O, pH: 6.8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, Department of Chemistry, University of Utah on 2016 Nov 11
|
The C-Terminal Zinc Fingers of ZBTB38 are Novel Selective Readers of DNA Methylation.
J Mol Biol 430(3):258-271 (2018)
Pozner A, Hudson NO, Trewhella J, Terooatea TW, Miller SA, Buck-Koehntop BA
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Mar 24
|
Characterizing Protein Dynamics with Integrative Use of Bulk and Single-Molecule Techniques.
Biochemistry 57(3):305-313 (2018)
Liu Z, Gong Z, Cao Y, Ding YH, Dong MQ, Lu YB, Zhang WP, Tang C
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
NHL repeat-containing protein 2 monomer, 80 kDa Homo sapiens protein
|
| Buffer: |
20 mM BisTris, 100 mM NaCl, 2 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jul 8
|
Structural analysis of human NHLRC2, mutations of which are associated with FINCA disease.
PLoS One 13(8):e0202391 (2018)
Biterova E, Ignatyev A, Uusimaa J, Hinttala R, Ruddock LW
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
121 |
nm3 |
|
|