|
|
|
|
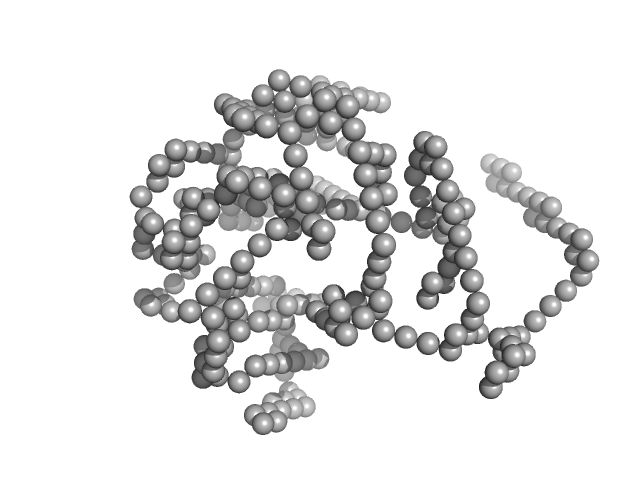
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.2 |
nm |
|
|
|
|
|
|
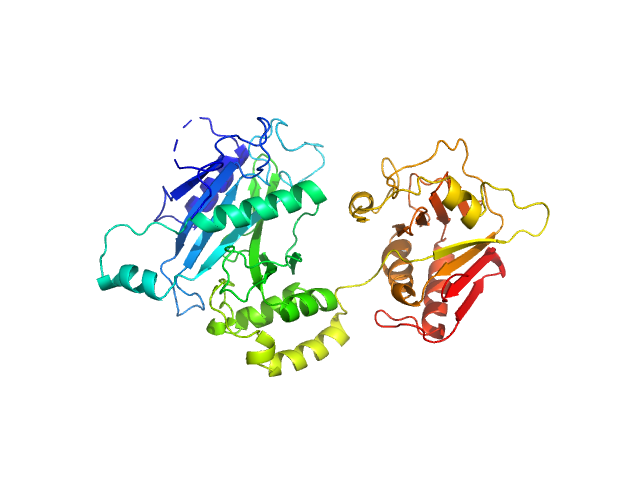
|
| Sample: |
Suppressor of fused homolog monomer, 53 kDa Drosophila melanogaster protein
|
| Buffer: |
50 mM bis-TRIS pH 5.5, 200 mM NaCl, 10% glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 May 7
|
Suppressor of Fused
Valerie Biou
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Putative DNA binding protein dimer, 37 kDa Bacillus subtilis subsp. … protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2016 Aug 17
|
YeeF, a polymorphic toxin from Bacillus subtilis, is a ribosomal RNase with promiscuous DNA binding property
Krishan Gopal
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
YezG, cognate immunity protein of YeeF monomer, 19 kDa Bacillus subtilis subsp. … protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2016 Aug 17
|
YeeF, a polymorphic toxin from Bacillus subtilis, is a ribosomal RNase with promiscuous DNA binding property
Krishan Gopal
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Putative DNA binding protein dimer, 37 kDa Bacillus subtilis subsp. … protein
YezG, cognate immunity protein of YeeF monomer, 19 kDa Bacillus subtilis subsp. … protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2016 Aug 18
|
YeeF, a polymorphic toxin from Bacillus subtilis, is a ribosomal RNase with promiscuous DNA binding property
Krishan Gopal
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
100 mM HEPES, 1 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 21
|
A high flux setup for millisecond-scale small-angle X-ray scattering studies on macromolecular solutions
Clement Blanchet
|
|
|
|
|
|
|
|
| Sample: |
PAS fold family dimer, 78 kDa Coleofasciculus chthonoplastes PCC … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 5 % w/v Glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at cSAXS, Swiss Light Source on 2015 Mar 11
|
MPAC Delta132
Robert Lindner
|
| RgGuinier |
4.4 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
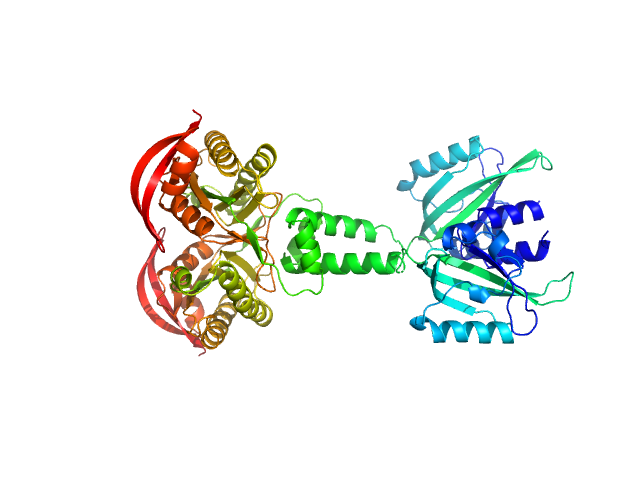
|
| Sample: |
PAS fold family dimer, 78 kDa Coleofasciculus chthonoplastes PCC … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 5 % w/v Glycerol, 1 mM ApCpp, pH: 7.5 |
| Experiment: |
SAXS
data collected at cSAXS, Swiss Light Source on 2015 Mar 11
|
MPAC Delta132
Robert Lindner
|
| RgGuinier |
4.0 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|
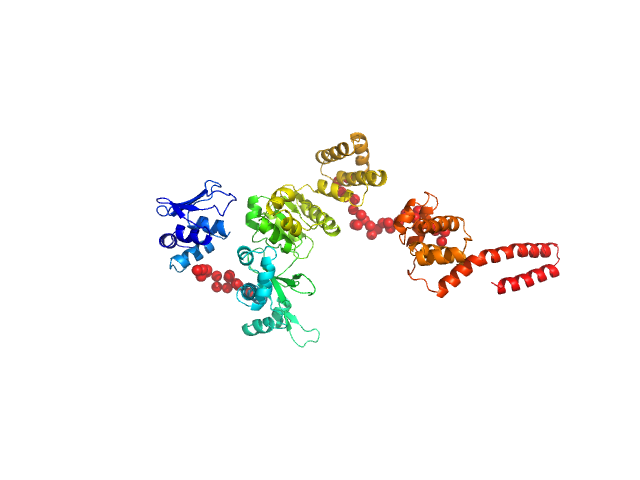
|
| Sample: |
DNA primase monomer, 69 kDa Bacillus subtilis protein
|
| Buffer: |
25 mM Tris-HCl,100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Oct 1
|
Full length of DnaG primase from Bacillus subtilis
Zhongchuan Liu
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
119 |
nm3 |
|
|
|
|
|
|
|
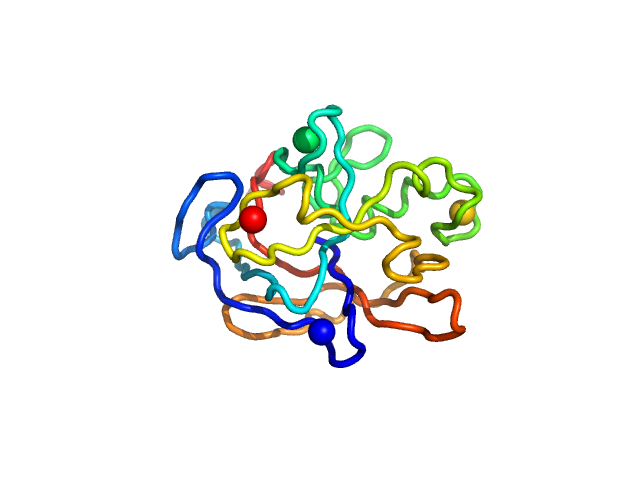
| Sample: |
Iron-regulated protein FrpC (amino acids 415-591) monomer, 19 kDa Neisseria meningitidis protein
|
| Buffer: |
5 mM Tris 50 mM NaCl 10 mM CaCl2 0.1% NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2013 Oct 3
|
Self-processing module of Iron-regulated protein FrpC
Vojtěch Kubáň
|
| RgGuinier |
1.7 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|