|
|
|
|
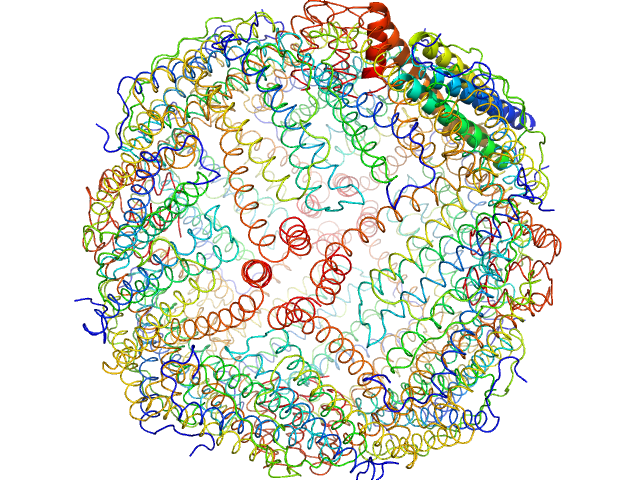
|
| Sample: |
Horse spleen apoferritin 24-mer, 476 kDa Equus caballus protein
|
| Buffer: |
tbs, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 15
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
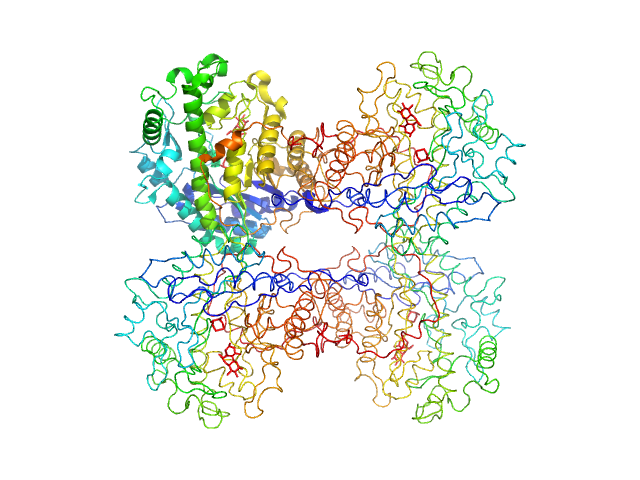
|
| Sample: |
Beta-amylase tetramer, 224 kDa Ipomoea batatas protein
|
| Buffer: |
tbs, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 15
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
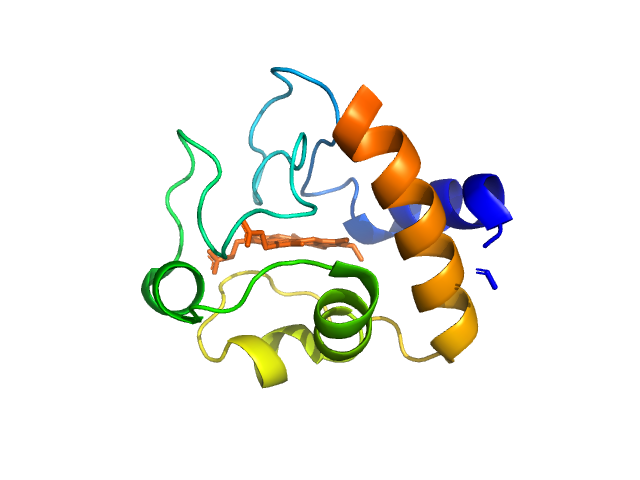
|
| Sample: |
Cytochrome c monomer, 12 kDa Equus caballus protein
|
| Buffer: |
tbs, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 16
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
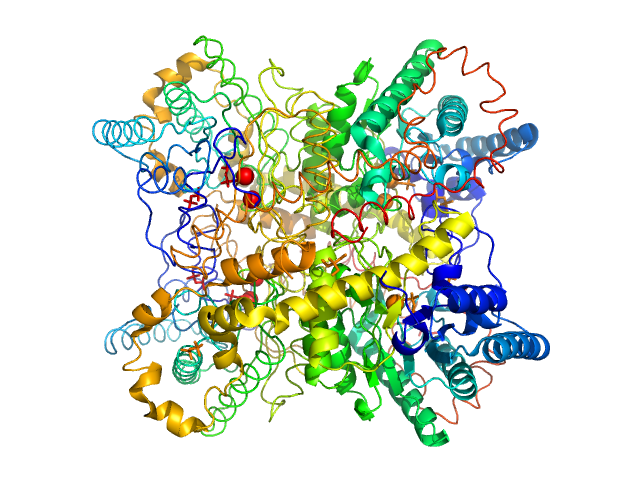
|
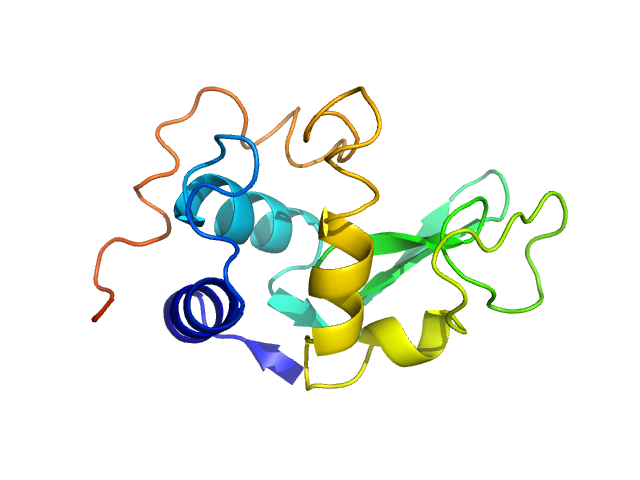
| Sample: |
Xylose isomerase tetramer, 172 kDa Streptomyces rubiginosus protein
|
| Buffer: |
100 mM tris pH 8.0, 100 mM NaCl, 1 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 17
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
|
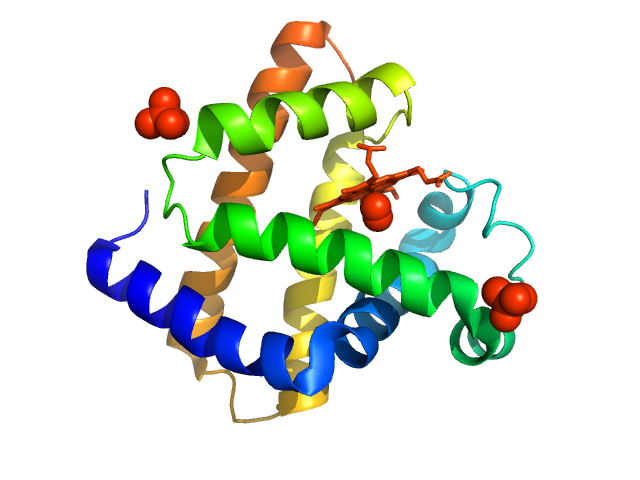
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM Sodium Acetate pH 4.0, 50 mM NaCl, pH: 4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 May 31
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
|
| Sample: |
Myoglobin monomer, 17 kDa Equus caballus protein
|
| Buffer: |
100 mM tris pH 7.5, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
WAXS benchmark on standard proteins
Maxim Petoukhov
|
|
|
|
|
|
|
|
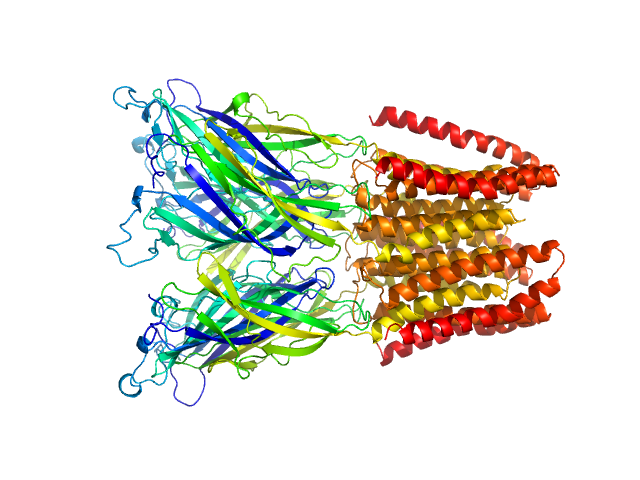
| Sample: |
Proton-gated ion channel pentamer, 183 kDa Gloeobacter violaceus (strain … protein
|
| Buffer: |
D2O, 20 mM Tris, 150 mM NaCl, 0.5 mM matched-out deuterated DDM,, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2019 Jun 20
|
Probing solution structure of the pentameric ligand-gated ion channel GLIC by small-angle neutron scattering
Proceedings of the National Academy of Sciences 118(37):e2108006118 (2021)
Lycksell M, Rovšnik U, Bergh C, Johansen N, Martel A, Porcar L, Arleth L, Howard R, Lindahl E
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
235 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Proton-gated ion channel pentamer, 183 kDa Gloeobacter violaceus (strain … protein
|
| Buffer: |
D2O, 20 mM Tris, 150 mM NaCl, 0.5 mM matched-out deuterated DDM,, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Aug 22
|
Probing solution structure of the pentameric ligand-gated ion channel GLIC by small-angle neutron scattering
Proceedings of the National Academy of Sciences 118(37):e2108006118 (2021)
Lycksell M, Rovšnik U, Bergh C, Johansen N, Martel A, Porcar L, Arleth L, Howard R, Lindahl E
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
|
|
|
|
|
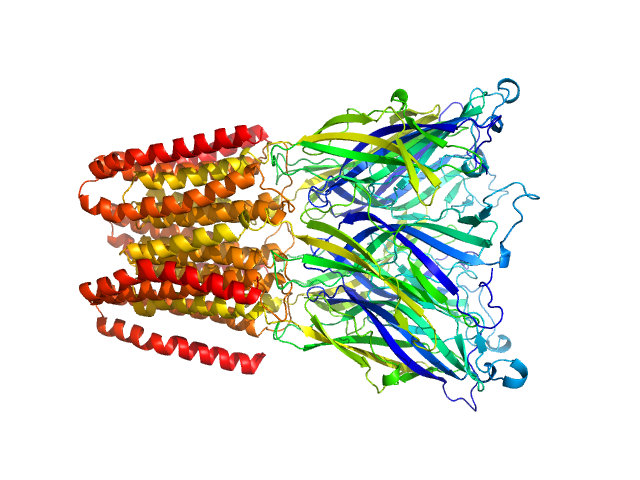
| Sample: |
Proton-gated ion channel pentamer, 183 kDa Gloeobacter violaceus (strain … protein
|
| Buffer: |
D2O, 20 mM Tris, 150 mM NaCl, 0.5 mM matched-out deuterated DDM,, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2019 Jun 21
|
Probing solution structure of the pentameric ligand-gated ion channel GLIC by small-angle neutron scattering
Proceedings of the National Academy of Sciences 118(37):e2108006118 (2021)
Lycksell M, Rovšnik U, Bergh C, Johansen N, Martel A, Porcar L, Arleth L, Howard R, Lindahl E
|
| RgGuinier |
4.0 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Proton-gated ion channel pentamer, 183 kDa Gloeobacter violaceus (strain … protein
|
| Buffer: |
D2O, 20 mM citrate, 150 mM NaCl, 0.5 mM match-out deuterated DDM, pH: 3 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Aug 22
|
Probing solution structure of the pentameric ligand-gated ion channel GLIC by small-angle neutron scattering
Proceedings of the National Academy of Sciences 118(37):e2108006118 (2021)
Lycksell M, Rovšnik U, Bergh C, Johansen N, Martel A, Porcar L, Arleth L, Howard R, Lindahl E
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
279 |
nm3 |
|
|