|
|
|
|
|
| Sample: |
Protein Gemin2 octamer, 230 kDa Drosophila melanogaster protein
Survival motor neuron protein octamer, 197 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Na/KPO4 pH 7.0, 300 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 19
|
Assembly of higher-order SMN oligomers is essential for metazoan viability and requires an exposed structural motif present in the YG zipper dimer.
Nucleic Acids Res 49(13):7644-7664 (2021)
Gupta K, Wen Y, Ninan NS, Raimer AC, Sharp R, Spring AM, Sarachan KL, Johnson MC, Van Duyne GD, Matera AG
|
| RgGuinier |
7.8 |
nm |
| Dmax |
28.7 |
nm |
| VolumePorod |
2660 |
nm3 |
|
|
|
|
|
|
|
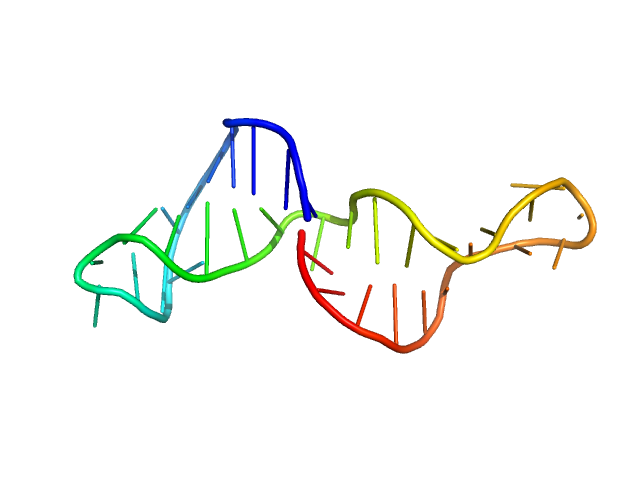
| Sample: |
Lung adenocarcinoma aptamer, truncated version monomer, 11 kDa Artificially synthesized DNA
|
| Buffer: |
Phosphate-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 28
|
The Role of Small-Angle X-Ray Scattering and Molecular Simulations in 3D Structure Elucidation of a DNA Aptamer Against Lung Cancer
Molecular Therapy - Nucleic Acids (2021)
Morozov D, Mironov V, Moryachkov R, Shchugoreva I, Artyushenko P, Zamay G, Kolovskaya O, Zamay T, Krat A, Molodenskiy D, Zabluda V, Veprintsev D, Sokolov A, Zukov R, Berezovski M, Tomilin F, Fedorov D, Alexeev Y, Kichkailo A
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
PSK, an antimicrobial peptide from Chrysomya megacephala monomer, 10 kDa Chrysomya megacephala protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Dec 7
|
Crystal and solution structures of a novel antimicrobial peptide from Chrysomya megacephala.
Acta Crystallogr D Struct Biol 77(Pt 7):894-903 (2021)
Xiao C, Xiao Z, Hu C, Lu J, Cui L, Zhang Y, Dai Y, Zhang Q, Wang S, Liu W
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Antitoxin ParD dodecamer, 108 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 18
|
Entropic pressure controls the oligomerization of the Vibrio cholerae
ParD2 antitoxin
Acta Crystallographica Section D Structural Biology 77(7):904-920 (2021)
Garcia-Rodriguez G, Girardin Y, Volkov A, Singh R, Muruganandam G, Van Dyck J, Sobott F, Versées W, Charlier D, Loris R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Myelin P2 disease mutant I50del monomer, 15 kDa protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jul 22
|
Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
FEBS J (2021)
Uusitalo M, Klenow MB, Laulumaa S, Blakeley MP, Simonsen AC, Ruskamo S, Kursula P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human P2 M114T mutant monomer, 15 kDa protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jul 22
|
Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
FEBS J (2021)
Uusitalo M, Klenow MB, Laulumaa S, Blakeley MP, Simonsen AC, Ruskamo S, Kursula P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human P2 V115A mutant monomer, 15 kDa protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jul 22
|
Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
FEBS J (2021)
Uusitalo M, Klenow MB, Laulumaa S, Blakeley MP, Simonsen AC, Ruskamo S, Kursula P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DciA monomer, 18 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Mar 26
|
Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase.
Nucleic Acids Res (2021)
Marsin S, Adam Y, Cargemel C, Andreani J, Baconnais S, Legrand P, Li de la Sierra-Gallay I, Humbert A, Aumont-Nicaise M, Velours C, Ochsenbein F, Durand D, Le Cam E, Walbott H, Possoz C, Quevillon-Cheruel S, Ferat JL
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Replicative DNA helicase (DnaB) hexamer, 317 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 1 mM ATP, pH: 8.8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Oct 4
|
Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase.
Nucleic Acids Res (2021)
Marsin S, Adam Y, Cargemel C, Andreani J, Baconnais S, Legrand P, Li de la Sierra-Gallay I, Humbert A, Aumont-Nicaise M, Velours C, Ochsenbein F, Durand D, Le Cam E, Walbott H, Possoz C, Quevillon-Cheruel S, Ferat JL
|
| RgGuinier |
4.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
585 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DnaB helicase complexed with ATP hexamer, 317 kDa Vibrio cholerae serotype … protein
DciA, 55 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 1 mM ATP, pH: 8.8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Oct 4
|
Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase.
Nucleic Acids Res (2021)
Marsin S, Adam Y, Cargemel C, Andreani J, Baconnais S, Legrand P, Li de la Sierra-Gallay I, Humbert A, Aumont-Nicaise M, Velours C, Ochsenbein F, Durand D, Le Cam E, Walbott H, Possoz C, Quevillon-Cheruel S, Ferat JL
|
| RgGuinier |
5.0 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
670 |
nm3 |
|
|