|
|
|
|
|
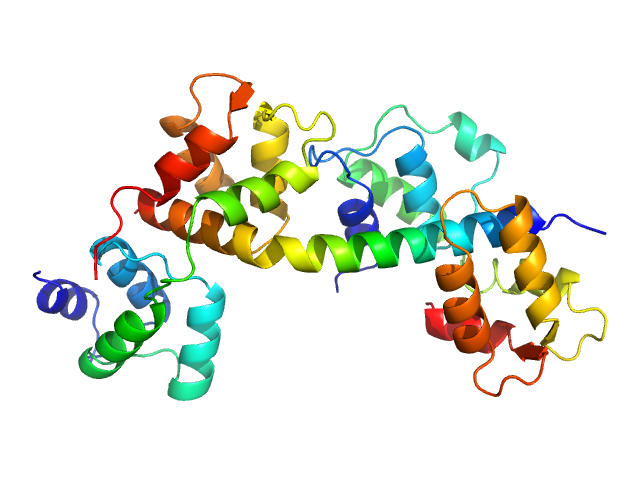
| Sample: |
Toxoplasma gondii essential light chain 1 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
Toxoplasma gondii myosin light chain, full length monomer, 24 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
...Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
69 |
nm3 |
|