|
|
|
|
|
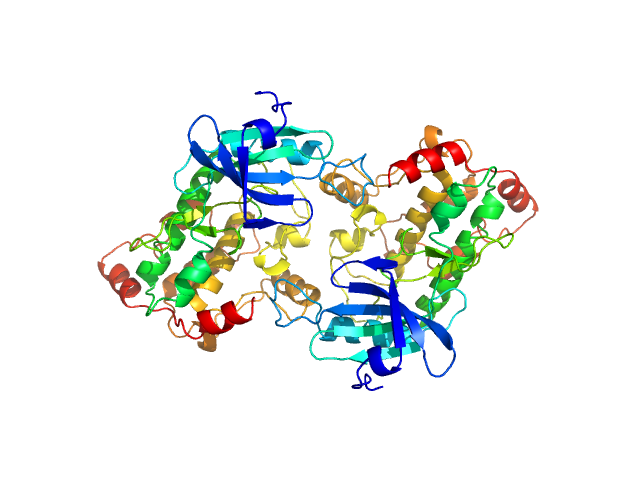
| Sample: |
Death associated protein kinase wild-type , 37 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 250 mM NaCl 5mM CaCl2 0.25 mM TCEP 5% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 18
|
Death-Associated Protein Kinase Activity Is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure 24(6):851-61 (2016)
...Kursula P, Schultz C, McCarthy AA, Hart DJ, Wilmanns M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
101 |
nm3 |
|