|
|
|
|
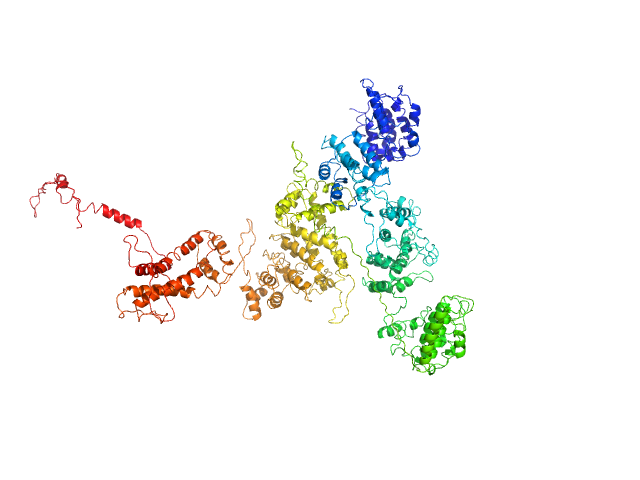
|
| Sample: |
Macrophage mannose receptor 1 dimer, 315 kDa Mouse myeloma cell … protein
|
| Buffer: |
50mM Hepes, 100mM NaCl, 1mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 15
|
Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses.
Sci Transl Med 12(530) (2020)
...Nguyen V, Bian Y, Zarif JC, de Groot AE, Mehta M, Fan L, Hu X, Simeonov A, Pate N, Abu-Asab M, Ferrer M, Southall N, Ock CY, Zhao Y, Lopez H, Kozlov S, de Val N, Yates CC, Baljinnyam B, Marugan J, Rud...
|
| RgGuinier |
7.9 |
nm |
| Dmax |
30.1 |
nm |
| VolumePorod |
584 |
nm3 |
|