|
|
|
|
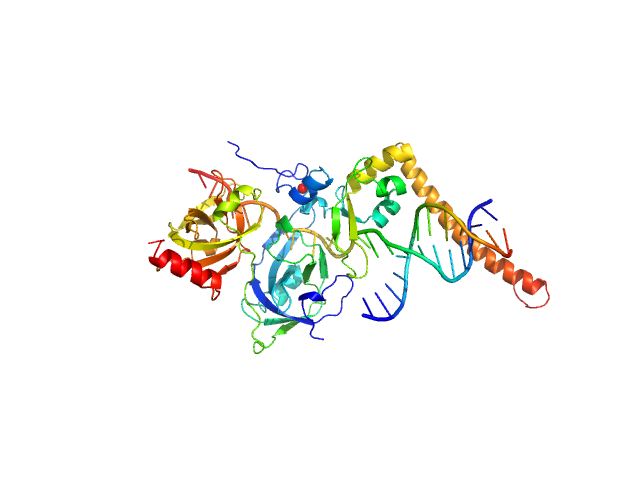
|
| Sample: |
DNA repair protein complementing XP-A cells monomer, 17 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 27 kDa Homo sapiens protein
3-prime Nucleotide Excision Repair Junction Model Substrate monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 17 Nov 2
|
A key interaction with RPA orients XPA in NER complexes.
Nucleic Acids Res (2020)
...Rosenberg D, Hammel M, Schärer OD, Chazin WJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
103 |
nm3 |
|