UniProt ID: Q14258 (189-630) E3 ubiquitin/ISG15 ligase TRIM25
UniProt ID: None (None-None) pre-let-7-a-1@1
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
Pre-let-7-a-1@1 monomer, 9 kDa synthetic construct RNA
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 4
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
5.7 |
nm |
| Dmax |
23.0 |
nm |
|
|
UniProt ID: Q14258 (189-630) E3 ubiquitin/ISG15 ligase TRIM25
UniProt ID: None (None-None) lnczc3h7a_304-326
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
Lnczc3h7a_304-326 monomer, 8 kDa Homo sapiens RNA
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 12
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
5.8 |
nm |
| Dmax |
16.6 |
nm |
|
|
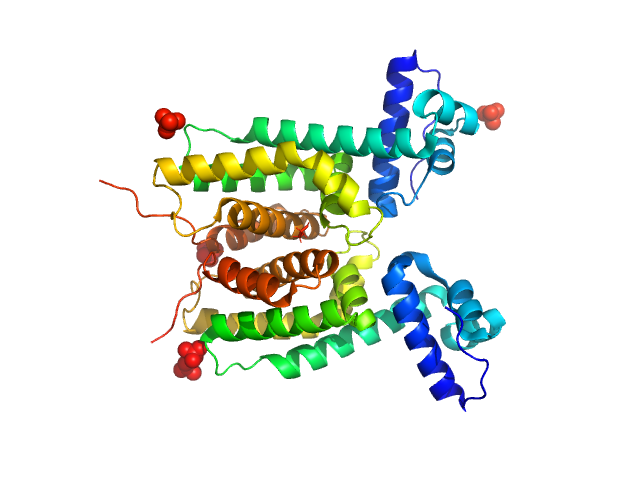
UniProt ID: Q9L8G8 (1-205) LuxR family transcriptional regulator
|
|
|
|
| Sample: |
LuxR family transcriptional regulator, 52 kDa Vibrio vulnificus protein
|
| Buffer: |
200 mM NaCl, 25 mM Tris, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 8
|
The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res 49(10):5967-5984 (2021)
Newman JD, Russell MM, Fan L, Wang YX, Gonzalez-Gutierrez G, van Kessel JC
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
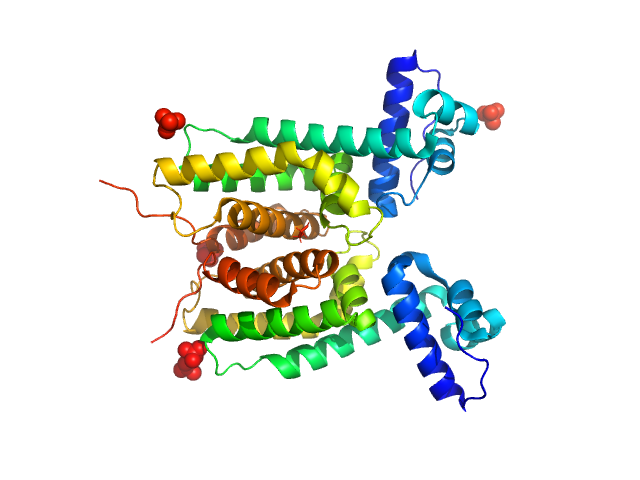
UniProt ID: Q9L8G8 (1-205) LuxR family transcriptional regulator, N55I
|
|
|
|
| Sample: |
LuxR family transcriptional regulator, N55I, 52 kDa Vibrio vulnificus protein
|
| Buffer: |
200 mM NaCl, 25 mM Tris, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Dec 11
|
The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res 49(10):5967-5984 (2021)
Newman JD, Russell MM, Fan L, Wang YX, Gonzalez-Gutierrez G, van Kessel JC
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
UniProt ID: Q9H5V8 (30-667) CUB domain-containing protein 1
|
|
|
|
| Sample: |
CUB domain-containing protein 1 monomer, 77 kDa Homo sapiens protein
|
| Buffer: |
10 mM phosphate, 137 mM NaCl, 2.7 mM KCl,, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 29
|
Targeting a proteolytic neoepitope on CUB domain containing protein 1 (CDCP1) for RAS-driven cancers.
J Clin Invest 132(4) (2022)
Lim SA, Zhou J, Martinko AJ, Wang YH, Filippova EV, Steri V, Wang D, Remesh SG, Liu J, Hann B, Kossiakoff AA, Evans MJ, Leung KK, Wells JA
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
185 |
nm3 |
|
|
UniProt ID: Q9H5V8 (30-367) Cleaved - CUB domain-containing protein 1 (N-terminus)
UniProt ID: Q9H5V8 (370-667) Cleaved - CUB domain-containing protein 1 (C-terminus)
|
|
|
|
| Sample: |
Cleaved - CUB domain-containing protein 1 (N-terminus) monomer, 38 kDa Homo sapiens protein
Cleaved - CUB domain-containing protein 1 (C-terminus) monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
10 mM phosphate, 137 mM NaCl, 2.7 mM KCl,, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 29
|
Targeting a proteolytic neoepitope on CUB domain containing protein 1 (CDCP1) for RAS-driven cancers.
J Clin Invest 132(4) (2022)
Lim SA, Zhou J, Martinko AJ, Wang YH, Filippova EV, Steri V, Wang D, Remesh SG, Liu J, Hann B, Kossiakoff AA, Evans MJ, Leung KK, Wells JA
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
182 |
nm3 |
|
|
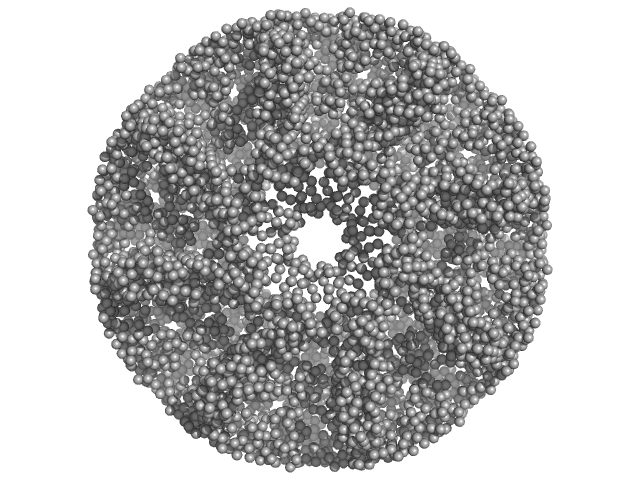
UniProt ID: Q9RVQ8 (27-500) Carboxypeptidase-related protein
|
|
|
|
| Sample: |
Carboxypeptidase-related protein 16-mer, 903 kDa Deinococcus radiodurans (strain … protein
|
| Buffer: |
20 mM Tris-Cl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2020 Dec 25
|
Novel oligomeric assembly of S10-carboxypeptidase from Deinococcus radiodurans
Dr. Rahul Singh
|
| RgGuinier |
7.4 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
1180 |
nm3 |
|
|
UniProt ID: P77399 (1-714) Fatty acid oxidation complex subunit alpha
|
|
|

|
| Sample: |
Fatty acid oxidation complex subunit alpha monomer, 77 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, 5% glycerol, 2.5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 9
|
Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial β-oxidation trifunctional enzymes.
Structure (2023)
Sah-Teli SK, Pinkas M, Hynönen MJ, Butcher SJ, Wierenga RK, Novacek J, Venkatesan R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
UniProt ID: P04275 (1596-1668) von Willebrand factor
|
|
|
|
| Sample: |
Von Willebrand factor monomer, 11 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 24
|
An Integrative Structural Biology Analysis of Von Willebrand Factor Binding and Processing by ADAMTS-13 in Solution.
J Mol Biol 433(13):166954 (2021)
Del Amo-Maestro L, Sagar A, Pompach P, Goulas T, Scavenius C, Ferrero DS, Castrillo-Briceño M, Taulés M, Enghild JJ, Bernadó P, Gomis-Rüth FX
|
| RgGuinier |
3.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: P04275 (1596-1668) von Willebrand factor
|
|
|
|
| Sample: |
Von Willebrand factor monomer, 11 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 24
|
An Integrative Structural Biology Analysis of Von Willebrand Factor Binding and Processing by ADAMTS-13 in Solution.
J Mol Biol 433(13):166954 (2021)
Del Amo-Maestro L, Sagar A, Pompach P, Goulas T, Scavenius C, Ferrero DS, Castrillo-Briceño M, Taulés M, Enghild JJ, Bernadó P, Gomis-Rüth FX
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
27 |
nm3 |
|
|