UniProt ID: P17846 (1-570) Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein)
UniProt ID: P38038 (53-599) Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein)
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein) monomer, 64 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2021 Apr 3
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.0 |
nm |
|
|
UniProt ID: P17846 (1-570) Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein)
UniProt ID: P38038 (53-599) Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein)
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein) monomer, 64 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2021 Apr 3
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.6 |
nm |
|
|
UniProt ID: P17846 (1-570) Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein)
UniProt ID: P38038 (53-599) Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein)
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein) monomer, 64 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2021 Apr 3
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.4 |
nm |
|
|
UniProt ID: P17846 (1-570) Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein)
UniProt ID: P38038 (53-599) Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein)
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein) monomer, 64 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2021 Apr 3
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.9 |
nm |
|
|
UniProt ID: P00698 (19-129) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
100 mM HEPES pH 7.5, 20 %(v/v) jeffamine M-600, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 28
|
Dependence of concentration of precursor clusters formed in lysozyme crystallization solutions on degree of supersaturation and its effect on character of solution transition from liquid to condensed phase
Petr Konarev
|
|
|
UniProt ID: P00698 (19-129) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
100 mM sodium acetate, pH 4.6, 2.0 M sodium formate, pH: 4.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 28
|
Dependence of concentration of precursor clusters formed in lysozyme crystallization solutions on degree of supersaturation and its effect on character of solution transition from liquid to condensed phase
Petr Konarev
|
|
|
UniProt ID: P00698 (19-129) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
200 mM K/Na tartrate, 100 mM tri-sodium citrate pH 5.6, 2.0 M ammonium sulfate, pH: 5.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 28
|
Dependence of concentration of precursor clusters formed in lysozyme crystallization solutions on degree of supersaturation and its effect on character of solution transition from liquid to condensed phase
Petr Konarev
|
|
|
UniProt ID: A0A160VQZ8 (1-267) Diacetylchitobiose deacetylase
|
|
|
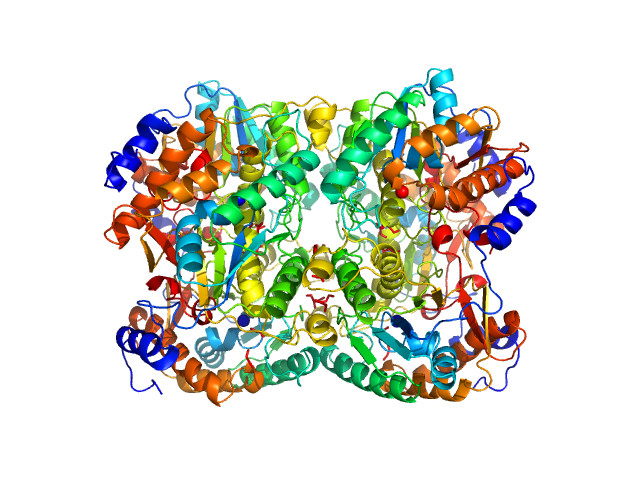
|
| Sample: |
Diacetylchitobiose deacetylase hexamer, 186 kDa Thermococcus chitonophagus protein
|
| Buffer: |
20 mM TRIS, 200 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Sep 26
|
Structural, Thermodynamic and Enzymatic Characterization of N,N-Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci 23(24) (2022)
Biniek-Antosiak K, Bejger M, Śliwiak J, Baranowski D, Mohammed ASA, Svergun DI, Rypniewski W
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
317 |
nm3 |
|
|
UniProt ID: A0A160VQZ8 (1-267) Diacetylchitobiose deacetylase
|
|
|
|
| Sample: |
Diacetylchitobiose deacetylase, 185 kDa Thermococcus chitonophagus protein
|
| Buffer: |
20 mM TRIS, 200 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Sep 26
|
Structural, Thermodynamic and Enzymatic Characterization of N,N-Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci 23(24) (2022)
Biniek-Antosiak K, Bejger M, Śliwiak J, Baranowski D, Mohammed ASA, Svergun DI, Rypniewski W
|
|
|
UniProt ID: P52003 (5-139) Multidrug resistance operon repressor
|
|
|

|
| Sample: |
Multidrug resistance operon repressor dimer, 32 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 10mM DTT, 1% v/v glycerol, pH: 7.1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 23
|
Small-angle X-ray and neutron scattering of MexR and its complex with DNA supports a conformational selection binding model
Biophysical Journal (2022)
Caporaletti F, Pietras Z, Morad V, Mårtensson L, Gabel F, Wallner B, Martel A, Sunnerhagen M
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
56 |
nm3 |
|
|