UniProt ID: P23246 (214-707) Splicing factor, proline- and glutamine-rich
|
|
|
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
|
| Buffer: |
150 mM KCl, 0.075% glycerol, 20 mM HEPES, 0.5 mM DTT,, pH: 7.4 |
| Experiment: |
SANS
data collected at Quokka - Small Angle Neutron Scattering, Australian Centre for Neutron Scattering (ANSTO) on 2023 Jan 20
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
6.1 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
UniProt ID: Q9SQQ6 (172-338) NAC domain-containing protein 46
|
|
|
|
| Sample: |
NAC domain-containing protein 46 monomer, 18 kDa Arabidopsis thaliana protein
|
| Buffer: |
20mM Na2HPO4/NaH2PO4, 100mM NaCl, 5mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Dec 1
|
Hierarchy in regulator interactions with distant transcriptional activation domains empowers rheostatic regulation
Protein Science 34(6) (2025)
Due A, Davey N, Thomasen F, Morffy N, Prestel A, Brakti I, O'Shea C, Strader L, Lindorff‐Larsen K, Skriver K, Kragelund B
|
| RgGuinier |
3.5 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
UniProt ID: Q9SQQ6 (172-338) NAC domain-containing protein 46
UniProt ID: Q8RY59 (499-572) Inactive poly [ADP-ribose] polymerase RCD1
|
|
|
|
| Sample: |
NAC domain-containing protein 46 monomer, 18 kDa Arabidopsis thaliana protein
Inactive poly [ADP-ribose] polymerase RCD1 monomer, 9 kDa Arabidopsis thaliana protein
|
| Buffer: |
20mM Na2HPO4/NaH2PO4, 100mM NaCl, 5mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Dec 1
|
Hierarchy in regulator interactions with distant transcriptional activation domains empowers rheostatic regulation
Protein Science 34(6) (2025)
Due A, Davey N, Thomasen F, Morffy N, Prestel A, Brakti I, O'Shea C, Strader L, Lindorff‐Larsen K, Skriver K, Kragelund B
|
| RgGuinier |
3.8 |
nm |
| Dmax |
20.6 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
UniProt ID: P48047 (None-None) ATP synthase subunit O, mitochondrial
|
|
|
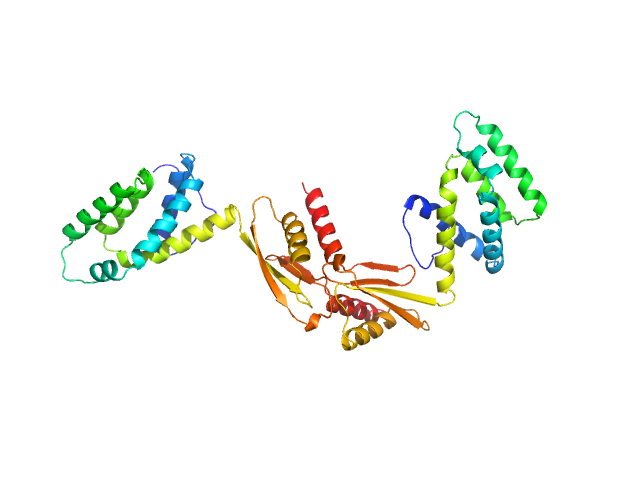
|
| Sample: |
ATP synthase subunit O, mitochondrial dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Mar 15
|
The OSCP Subunit of ATP Synthase is a Dimer in Solution: Strategy to Induce the Monomeric Protein as a New Tool for Drug Discovery.
J Mol Biol 437(17):169267 (2025)
Fabbian S, Gabbatore L, Morbiato L, Zotti M, Giorgio V, Battisutta R, Giachin G, Sosic A, Bellanda M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
UniProt ID: P48047 (137-213) ATP synthase subunit O, mitochondrial C-terminal section
|
|
|
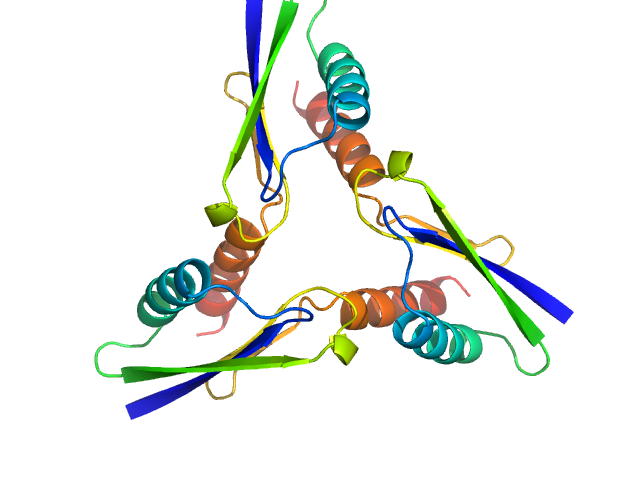
|
| Sample: |
ATP synthase subunit O, mitochondrial C-terminal section trimer, 25 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Mar 15
|
The OSCP Subunit of ATP Synthase is a Dimer in Solution: Strategy to Induce the Monomeric Protein as a New Tool for Drug Discovery.
J Mol Biol 437(17):169267 (2025)
Fabbian S, Gabbatore L, Morbiato L, Zotti M, Giorgio V, Battisutta R, Giachin G, Sosic A, Bellanda M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: Q8A9M2 (1-438) Xylose isomerase
|
|
|

|
| Sample: |
Xylose isomerase tetramer, 196 kDa Bacteroides thetaiotaomicron (strain … protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Oct 22
|
BioSAXS Australian Synchrotron Standard Protein
Annmaree Warrender
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
222 |
nm3 |
|
|
UniProt ID: Q9KLD5 (24-485) GlcNAc-binding protein A
|
|
|
|
| Sample: |
GlcNAc-binding protein A monomer, 51 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
100 mM NaCl, 20 mM Tris, 47% v/v D₂O, pH: 8 |
| Experiment: |
SANS
data collected at D11, ILL on 2020 Aug 18
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
Henrik Vinther Sørensen
|
|
|
UniProt ID: Q9KLD5 (24-485) GlcNAc-binding protein A (perdeuterated)
|
|
|
|
| Sample: |
GlcNAc-binding protein A (perdeuterated) monomer, 51 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
100 mM NaCl, 20 mM Tris, 47% v/v D₂O, pH: 8 |
| Experiment: |
SANS
data collected at D11, ILL on 2019 Sep 21
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
Henrik Vinther Sørensen
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.7 |
nm |
|
|
UniProt ID: None (None-None) β-chitin nanofibers from squid pens
UniProt ID: Q9KLD5 (24-485) GlcNAc-binding protein A (perdeuterated)
|
|
|
|
| Sample: |
Β-chitin nanofibers from squid pens None, Squid
GlcNAc-binding protein A (perdeuterated) monomer, 51 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM acetate, 47% v/v D₂O, pH: 5 |
| Experiment: |
SANS
data collected at D11, ILL on 2020 Aug 18
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
Henrik Vinther Sørensen
|
|
|
UniProt ID: None (None-None) β-chitin nanofibers from squid pens
UniProt ID: Q9KLD5 (24-485) GlcNAc-binding protein A (perdeuterated)
|
|
|
|
| Sample: |
Β-chitin nanofibers from squid pens None, Squid
GlcNAc-binding protein A (perdeuterated) monomer, 51 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM acetate, 47% v/v D₂O, pH: 5 |
| Experiment: |
SANS
data collected at D11, ILL on 2020 Aug 18
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
Henrik Vinther Sørensen
|
|
|