UniProt ID: Q6PHU5 (None-None) Sortilin 1 A464E alias Neurotensin-receptor 3 A464E
|
|
|

|
| Sample: |
Sortilin 1 A464E alias Neurotensin-receptor 3 A464E monomer, 76 kDa Mus musculus protein
|
| Buffer: |
25 mM MES pH 5.5, 150 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
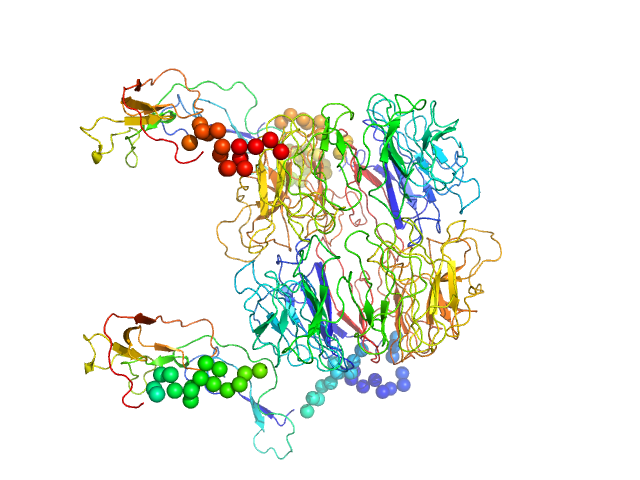
UniProt ID: Q6PHU5 (None-None) Sortilin, also: Neurotensin-receptor 3
|
|
|
|
| Sample: |
Sortilin, also: Neurotensin-receptor 3 dimer, 153 kDa Mus musculus protein
|
| Buffer: |
25 mM MES pH 5.5, 150 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
336 |
nm3 |
|
|
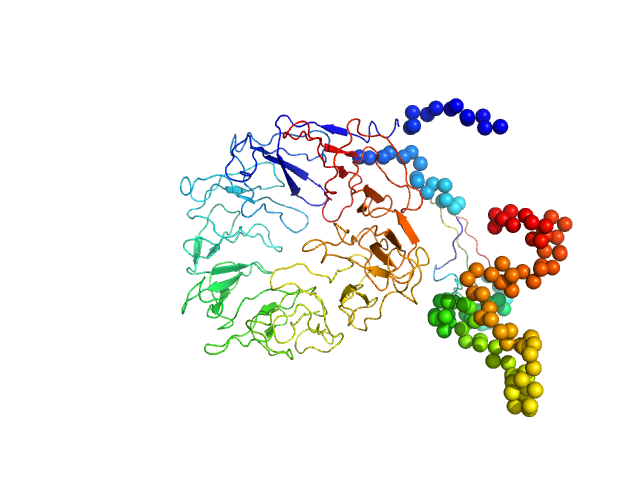
UniProt ID: Q6PHU5 (None-None) Sortilin 1 A464E alias Neurotensin-receptor 3 A464E
|
|
|
|
| Sample: |
Sortilin 1 A464E alias Neurotensin-receptor 3 A464E monomer, 76 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
UniProt ID: P11274 (None-None) BCR-ABL p210 fusion protein (DH-PH)
|
|
|
|
| Sample: |
BCR-ABL p210 fusion protein (DH-PH) monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% Glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 13
|
Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun 8(1):2101 (2017)
Reckel S, Gehin C, Tardivon D, Georgeon S, Kükenshöner T, Löhr F, Koide A, Buchner L, Panjkovich A, Reynaud A, Pinho S, Gerig B, Svergun D, Pojer F, Güntert P, Dötsch V, Koide S, Gavin AC, Hantschel O
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
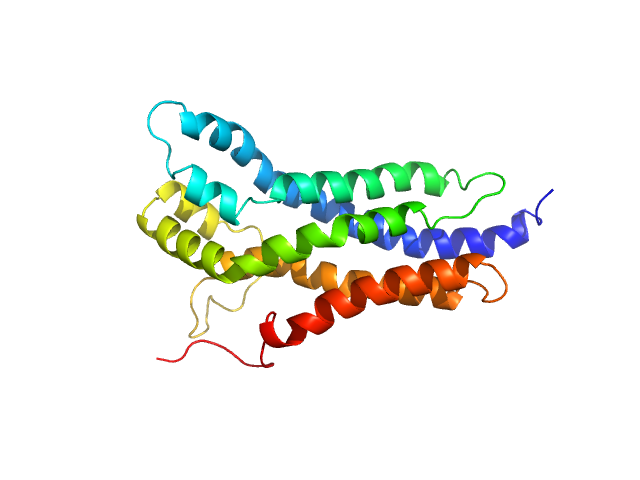
UniProt ID: P11274 (None-None) BCR-ABL p210 fusion protein
|
|
|
|
| Sample: |
BCR-ABL p210 fusion protein monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% Glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 13
|
Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun 8(1):2101 (2017)
Reckel S, Gehin C, Tardivon D, Georgeon S, Kükenshöner T, Löhr F, Koide A, Buchner L, Panjkovich A, Reynaud A, Pinho S, Gerig B, Svergun D, Pojer F, Güntert P, Dötsch V, Koide S, Gavin AC, Hantschel O
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
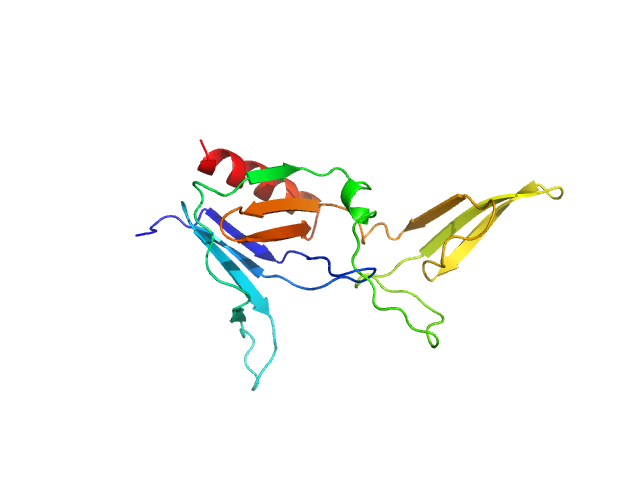
UniProt ID: P11274 (None-None) BCR-ABL p210 fusion protein (PH domain)
|
|
|
|
| Sample: |
BCR-ABL p210 fusion protein (PH domain) monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% Glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Nov 10
|
Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun 8(1):2101 (2017)
Reckel S, Gehin C, Tardivon D, Georgeon S, Kükenshöner T, Löhr F, Koide A, Buchner L, Panjkovich A, Reynaud A, Pinho S, Gerig B, Svergun D, Pojer F, Güntert P, Dötsch V, Koide S, Gavin AC, Hantschel O
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
UniProt ID: O34627 (2-261) pfyP - Blue light photoreceptor
|
|
|

|
| Sample: |
PfyP - Blue light photoreceptor dimer, 58 kDa Bacillus subtilis protein
|
| Buffer: |
PBS + 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Oct 24
|
The switch that does not flip: the blue-light receptor YtvA from Bacillus subtilis adopts an elongated dimer conformation independent of the activation state as revealed by a combined AUC and SAXS study.
J Mol Biol 403(1):78-87 (2010)
Jurk M, Dorn M, Kikhney A, Svergun D, Gärtner W, Schmieder P
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
UniProt ID: O34627 (2-261) pfyP - Blue light photoreceptor
|
|
|

|
| Sample: |
PfyP - Blue light photoreceptor dimer, 58 kDa Bacillus subtilis protein
|
| Buffer: |
PBS + 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Oct 24
|
The switch that does not flip: the blue-light receptor YtvA from Bacillus subtilis adopts an elongated dimer conformation independent of the activation state as revealed by a combined AUC and SAXS study.
J Mol Biol 403(1):78-87 (2010)
Jurk M, Dorn M, Kikhney A, Svergun D, Gärtner W, Schmieder P
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
UniProt ID: P20273 (None-None) CD22 extracellular domain
|
|
|

|
| Sample: |
CD22 extracellular domain monomer, 90 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 21
|
Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun 8(1):764 (2017)
Ereño-Orbea J, Sicard T, Cui H, Mazhab-Jafari MT, Benlekbir S, Guarné A, Rubinstein JL, Julien JP
|
| RgGuinier |
8.0 |
nm |
| Dmax |
30.6 |
nm |
|
|
UniProt ID: P20273 (None-None) CD22 extracellular domain
UniProt ID: (None-None) alpha(2,6)-Sialyllactose
|
|
|
|
| Sample: |
CD22 extracellular domain monomer, 90 kDa Homo sapiens protein
Alpha(2,6)-Sialyllactose, 1 kDa
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 21
|
Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun 8(1):764 (2017)
Ereño-Orbea J, Sicard T, Cui H, Mazhab-Jafari MT, Benlekbir S, Guarné A, Rubinstein JL, Julien JP
|
| RgGuinier |
8.1 |
nm |
| Dmax |
29.8 |
nm |
|
|