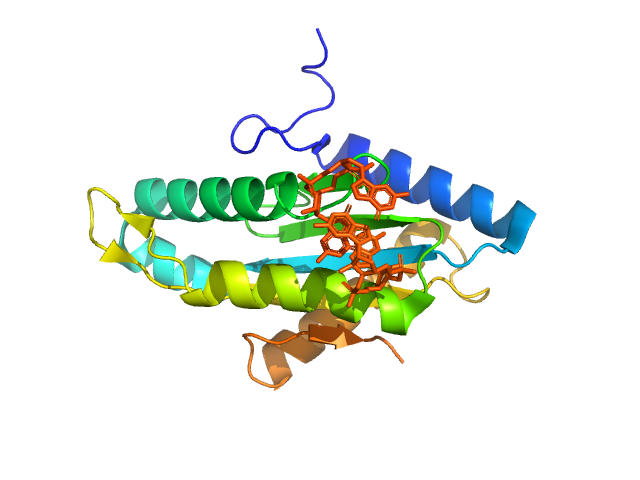
UniProt ID: A1U3W3 (None-None) Diguanylate cyclase
|
|
|
|
| Sample: |
Diguanylate cyclase monomer, 20 kDa Marinobacter hydrocarbonoclasticus protein
|
| Buffer: |
5 mM DTT 100 mM NaCl 10 mM Tris-HCl 0.02 % NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2010 Feb 12
|
Small angle X-ray scattering as a complementary tool for high-throughput structural studies.
Biopolymers 95(8):517-30 (2011)
Grant TD, Luft JR, Wolfley JR, Tsuruta H, Martel A, Montelione GT, Snell EH
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
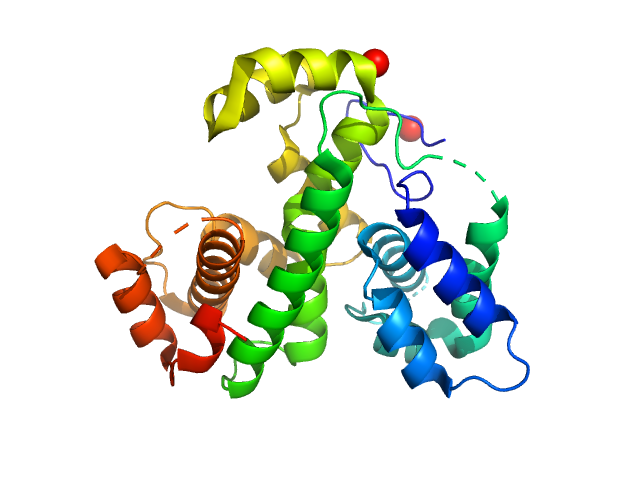
UniProt ID: Q7WZ31 (None-None) MmoQ
|
|
|
|
| Sample: |
MmoQ monomer, 32 kDa Methylococcus capsulatus protein
|
| Buffer: |
5 mM DTT 100 mM NaCl 10 mM Tris-HCl 0.02 % NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2010 Feb 12
|
Small angle X-ray scattering as a complementary tool for high-throughput structural studies.
Biopolymers 95(8):517-30 (2011)
Grant TD, Luft JR, Wolfley JR, Tsuruta H, Martel A, Montelione GT, Snell EH
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
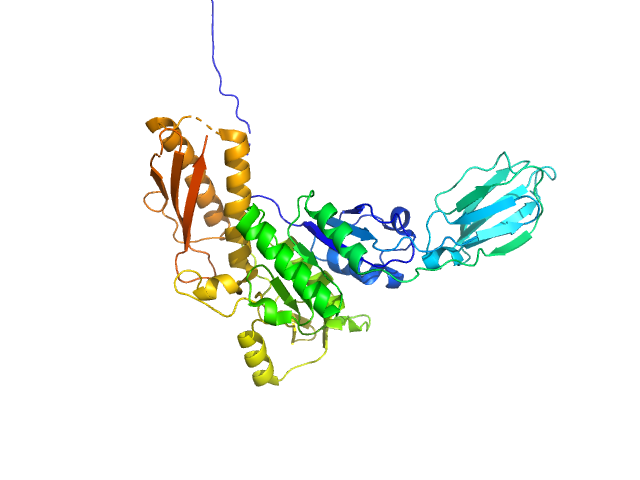
UniProt ID: Q24QE6 (None-None) Uncharacterized protein
|
|
|
|
| Sample: |
Uncharacterized protein monomer, 48 kDa Desulfitobacterium hafniense protein
|
| Buffer: |
5 mM DTT 100 mM NaCl 10 mM Tris-HCl 0.02 % NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2010 Feb 12
|
Small angle X-ray scattering as a complementary tool for high-throughput structural studies.
Biopolymers 95(8):517-30 (2011)
Grant TD, Luft JR, Wolfley JR, Tsuruta H, Martel A, Montelione GT, Snell EH
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
UniProt ID: P0CG48 (305-379) Polyubiquitin-C
|
|
|
|
| Sample: |
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
|
| Buffer: |
100mM NaCl, 10mM sodium acetate, pH: 6 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Mar 24
|
Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition.
Elife 4 (2015)
Liu Z, Gong Z, Jiang WX, Yang J, Zhu WK, Guo DC, Zhang WP, Liu ML, Tang C
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
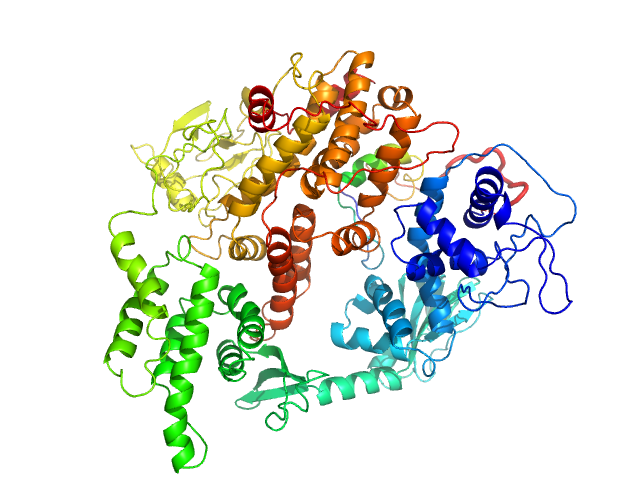
UniProt ID: O95398-3 (2-881) Rap guanine nucleotide exchange factor 3
|
|
|
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 100 kDa Homo sapiens protein
|
| Buffer: |
1mM EDTA, 10mM DTT, 500mM NaCl, and 10mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2012 Sep 7
|
Conformational States of Exchange Protein Directly Activated by cAMP (EPAC1) Revealed by Ensemble Modeling and Integrative Structural Biology.
Cells 9(1) (2019)
White MA, Tsalkova T, Mei FC, Cheng X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
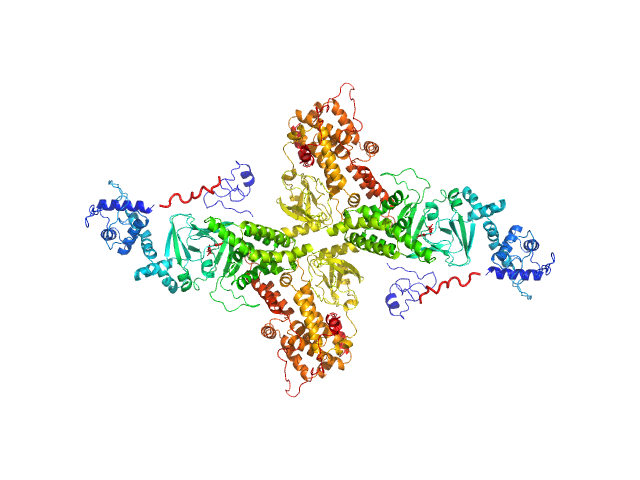
UniProt ID: O95398-3 (2-881) Rap guanine nucleotide exchange factor 3 (dimer)
|
|
|
|
| Sample: |
Rap guanine nucleotide exchange factor 3 (dimer) dimer, 200 kDa Homo sapiens protein
|
| Buffer: |
1mM EDTA, 10mM DTT, 500mM NaCl, 1mM cAMP, and 10mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2012 Jan 30
|
Conformational States of Exchange Protein Directly Activated by cAMP (EPAC1) Revealed by Ensemble Modeling and Integrative Structural Biology.
Cells 9(1) (2019)
White MA, Tsalkova T, Mei FC, Cheng X
|
| RgGuinier |
5.3 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
415 |
nm3 |
|
|
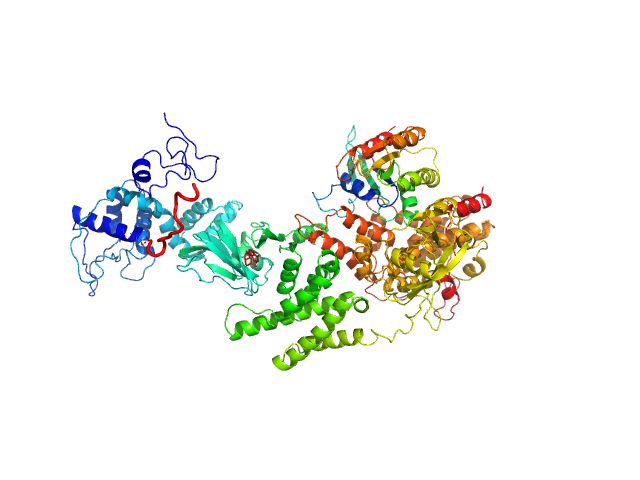
UniProt ID: O95398-3 (2-881) Rap guanine nucleotide exchange factor 3
UniProt ID: Q52L50 (1-167) RAS related protein 1b
|
|
|
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 100 kDa Homo sapiens protein
RAS related protein 1b monomer, 18 kDa Mus musculus protein
|
| Buffer: |
1mM EDTA, 10mM DTT, 500mM NaCl, 1mM cAMP, and 10mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2013 Apr 1
|
Conformational States of Exchange Protein Directly Activated by cAMP (EPAC1) Revealed by Ensemble Modeling and Integrative Structural Biology.
Cells 9(1) (2019)
White MA, Tsalkova T, Mei FC, Cheng X
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
UniProt ID: (None-None) 169 bp DNA (145 bp Widom 601, flanked by 12bp DNA)
UniProt ID: P06897 (None-None) Histone H2A type 1
UniProt ID: P02281 (None-None) Histone H2B 1.1
UniProt ID: P84233 (1-136) Histone H3.2
UniProt ID: P62799 (None-None) Histone H4
|
|
|
|
| Sample: |
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, ADP-BeF3 (0.5 mM ADP, 4 mM NaF, 0.6 mM BeCl2), pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.0 |
nm |
|
|
UniProt ID: P32657 (118-1274) Chromodomain-helicase-DNA-binding protein 1
UniProt ID: (None-None) 169 bp DNA (145 bp Widom 601, flanked by 12bp DNA)
UniProt ID: P06897 (None-None) Histone H2A type 1
UniProt ID: P02281 (None-None) Histone H2B 1.1
UniProt ID: P84233 (1-136) Histone H3.2
UniProt ID: P62799 (None-None) Histone H4
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
12.8 |
nm |
|
|
UniProt ID: P32657 (118-1274) Chromodomain-helicase-DNA-binding protein 1
UniProt ID: (None-None) 169 bp DNA (145 bp Widom 601, flanked by 12bp DNA)
UniProt ID: P06897 (None-None) Histone H2A type 1
UniProt ID: P02281 (None-None) Histone H2B 1.1
UniProt ID: P84233 (1-136) Histone H3.2
UniProt ID: P62799 (None-None) Histone H4
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, ADP-BeF3 (0.5 mM ADP, 4 mM NaF, 0.6 mM BeCl2), pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.5 |
nm |
|
|