UniProt ID: B4VKN6 (133-483) PAS fold family
|
|
|
|
| Sample: |
PAS fold family dimer, 78 kDa Coleofasciculus chthonoplastes PCC … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 5 % w/v Glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at cSAXS, Swiss Light Source on 2015 Mar 11
|
MPAC Delta132
Robert Lindner
|
| RgGuinier |
4.4 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
UniProt ID: B4VKN6 (133-483) PAS fold family
|
|
|
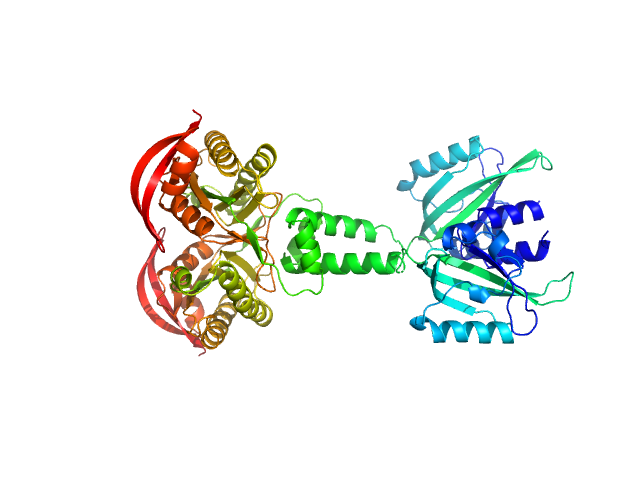
|
| Sample: |
PAS fold family dimer, 78 kDa Coleofasciculus chthonoplastes PCC … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 5 % w/v Glycerol, 1 mM ApCpp, pH: 7.5 |
| Experiment: |
SAXS
data collected at cSAXS, Swiss Light Source on 2015 Mar 11
|
MPAC Delta132
Robert Lindner
|
| RgGuinier |
4.0 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
UniProt ID: Q99717 (9-138) Mothers against decapentaplegic homolog 5
|
|
|
|
| Sample: |
Mothers against decapentaplegic homolog 5, 15 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 14
|
Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
(2019)
Ruiz L, Kaczmarska Z, Gomes T, Aragón E, Torner C, Freier R, Bagiński B, Martin-Malpartida P, de Martin Garrido N, Márquez J, Cordeiro T, Pluta R, Macias M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: O15198 (16-141) Mothers against decapentaplegic homolog 8_9
|
|
|
|
| Sample: |
Mothers against decapentaplegic homolog 8_9, 15 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 1
|
Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
(2019)
Ruiz L, Kaczmarska Z, Gomes T, Aragón E, Torner C, Freier R, Bagiński B, Martin-Malpartida P, de Martin Garrido N, Márquez J, Cordeiro T, Pluta R, Macias M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
UniProt ID: P21513 (1-529) Endoribonuclease E
|
|
|

|
| Sample: |
Endoribonuclease E tetramer, 247 kDa Escherichia coli protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
468 |
nm3 |
|
|
UniProt ID: Q74TC3 (1-529) Endoribonuclease E
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 248 kDa Yersinia pestis protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
470 |
nm3 |
|
|
UniProt ID: Q5NFK7 (1-543) Endoribonuclease E
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 256 kDa Francisella tularensis protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
491 |
nm3 |
|
|
UniProt ID: A0A0H3HN63 (1-532) Endoribonuclease E
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 250 kDa Burkholderia pseudomallei protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
437 |
nm3 |
|
|
UniProt ID: A0A0B9WR03 (1-544) Endoribonuclease E
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 254 kDa Acinetobacter baumannii protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
508 |
nm3 |
|
|
UniProt ID: P9WMU1 (None-None) Cell wall synthesis protein Wag31
|
|
|
|
| Sample: |
Cell wall synthesis protein Wag31, 30 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50mM Tris pH7.5, 300mM NaCl, 10% Glycerol, 1mM EDTA (ethylene diamine tetra acetic acid), 5mM β-mercaptoethanol (BME), pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 15
|
Higher order assembling of the mycobacterial polar growth factor DivIVA/Wag31.
J Struct Biol :107429 (2019)
Choukate K, Gupta A, Basu B, Virk K, Ganguli M, Chaudhuri B
|
| RgGuinier |
19.8 |
nm |
| Dmax |
23.4 |
nm |
|
|