UniProt ID: P9WMU1 (None-None) Cell wall synthesis protein Wag31
|
|
|
|
| Sample: |
Cell wall synthesis protein Wag31, 30 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50mM Tris pH7.5, 300mM NaCl, 10% Glycerol, 1mM EDTA (ethylene diamine tetra acetic acid), 5mM β-mercaptoethanol (BME), pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 15
|
Higher order assembling of the mycobacterial polar growth factor DivIVA/Wag31.
J Struct Biol :107429 (2019)
Choukate K, Gupta A, Basu B, Virk K, Ganguli M, Chaudhuri B
|
| RgGuinier |
21.8 |
nm |
| Dmax |
13.6 |
nm |
|
|
UniProt ID: O60828 (1-265) Polyglutamine-binding protein 1
|
|
|
|
| Sample: |
Polyglutamine-binding protein 1 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1mM DTT,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 18
|
Solution model of the intrinsically disordered polyglutamine tract-binding protein-1.
Biophys J 102(7):1608-16 (2012)
Rees M, Gorba C, de Chiara C, Bui TT, Garcia-Maya M, Drake AF, Okazawa H, Pastore A, Svergun D, Chen YW
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
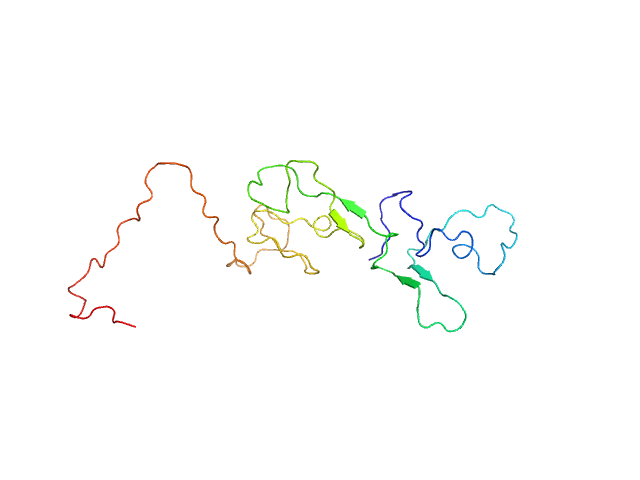
UniProt ID: P03372 (1-184) Estrogen receptor
|
|
|
|
| Sample: |
Estrogen receptor monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 0.05 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2017 Jun 12
|
A Metastable Contact and Structural Disorder in the Estrogen Receptor Transactivation Domain.
Structure 27(2):229-240.e4 (2019)
Peng Y, Cao S, Kiselar J, Xiao X, Du Z, Hsieh A, Ko S, Chen Y, Agrawal P, Zheng W, Shi W, Jiang W, Yang L, Chance MR, Surewicz WK, Buck M, Yang S
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
|
|
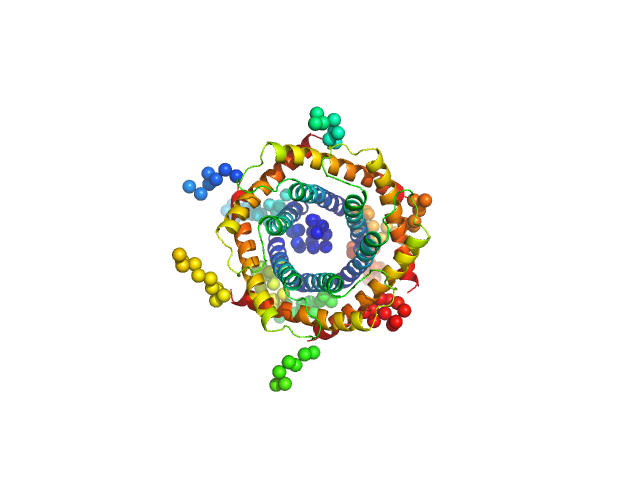
UniProt ID: O74700 (None-None) Mitochondrial import inner membrane translocase subunit TIM9
UniProt ID: P87108 (None-None) Mitochondrial import inner membrane translocase subunit TIM10
|
|
|
|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM9 trimer, 31 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM10 trimer, 31 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Tris, 150mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 27
|
Structural Basis of Membrane Protein Chaperoning through the Mitochondrial Intermembrane Space.
Cell 175(5):1365-1379.e25 (2018)
Weinhäupl K, Lindau C, Hessel A, Wang Y, Schütze C, Jores T, Melchionda L, Schönfisch B, Kalbacher H, Bersch B, Rapaport D, Brennich M, Lindorff-Larsen K, Wiedemann N, Schanda P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
UniProt ID: O74700 (None-None) Mitochondrial import inner membrane translocase subunit TIM9
UniProt ID: P87108 (None-None) Mitochondrial import inner membrane translocase subunit TIM10
UniProt ID: P38988 (8-300) Mitochondrial GTP/GDP carrier protein 1
|
|
|

|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM9 hexamer, 61 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM10 hexamer, 61 kDa Saccharomyces cerevisiae protein
Mitochondrial GTP/GDP carrier protein 1 monomer, 33 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Tris, 150mM NaCl, imidiazole, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 22
|
Structural Basis of Membrane Protein Chaperoning through the Mitochondrial Intermembrane Space.
Cell 175(5):1365-1379.e25 (2018)
Weinhäupl K, Lindau C, Hessel A, Wang Y, Schütze C, Jores T, Melchionda L, Schönfisch B, Kalbacher H, Bersch B, Rapaport D, Brennich M, Lindorff-Larsen K, Wiedemann N, Schanda P
|
| RgGuinier |
4.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
272 |
nm3 |
|
|
UniProt ID: Q02199 (121-154) Nucleoporin NUP49/NSP49
|
|
|
|
| Sample: |
Nucleoporin NUP49/NSP49 monomer, 4 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 8
|
Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements.
Proc Natl Acad Sci U S A 114(31):E6342-E6351 (2017)
Fuertes G, Banterle N, Ruff KM, Chowdhury A, Mercadante D, Koehler C, Kachala M, Estrada Girona G, Milles S, Mishra A, Onck PR, Gräter F, Esteban-Martín S, Pappu RV, Svergun DI, Lemke EA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
UniProt ID: Q02199 (121-154) Nucleoporin NUP49/NSP49
UniProt ID: None (None-None) Alexa Fluor™ 594 C5 Maleimide
UniProt ID: None (None-None) Alexa Fluor™ 488 C5 Hydroxylamine
|
|
|
|
| Sample: |
Nucleoporin NUP49/NSP49 monomer, 4 kDa Saccharomyces cerevisiae protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 8
|
Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements.
Proc Natl Acad Sci U S A 114(31):E6342-E6351 (2017)
Fuertes G, Banterle N, Ruff KM, Chowdhury A, Mercadante D, Koehler C, Kachala M, Estrada Girona G, Milles S, Mishra A, Onck PR, Gräter F, Esteban-Martín S, Pappu RV, Svergun DI, Lemke EA
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
UniProt ID: Q02199 (121-154) Nucleoporin NUP49/NSP49
|
|
|
|
| Sample: |
Nucleoporin NUP49/NSP49 monomer, 4 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 8
|
Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements.
Proc Natl Acad Sci U S A 114(31):E6342-E6351 (2017)
Fuertes G, Banterle N, Ruff KM, Chowdhury A, Mercadante D, Koehler C, Kachala M, Estrada Girona G, Milles S, Mishra A, Onck PR, Gräter F, Esteban-Martín S, Pappu RV, Svergun DI, Lemke EA
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
UniProt ID: Q02199 (121-154) Nucleoporin NUP49/NSP49
UniProt ID: None (None-None) Alexa Fluor™ 594 C5 Maleimide
UniProt ID: None (None-None) Alexa Fluor™ 488 C5 Hydroxylamine
|
|
|
|
| Sample: |
Nucleoporin NUP49/NSP49 monomer, 4 kDa Saccharomyces cerevisiae protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 8
|
Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements.
Proc Natl Acad Sci U S A 114(31):E6342-E6351 (2017)
Fuertes G, Banterle N, Ruff KM, Chowdhury A, Mercadante D, Koehler C, Kachala M, Estrada Girona G, Milles S, Mishra A, Onck PR, Gräter F, Esteban-Martín S, Pappu RV, Svergun DI, Lemke EA
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
7 |
nm3 |
|
|
UniProt ID: Q03281 (99-140) Inner nuclear membrane protein HEH2
|
|
|
|
| Sample: |
Inner nuclear membrane protein HEH2 monomer, 5 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 25
|
Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements.
Proc Natl Acad Sci U S A 114(31):E6342-E6351 (2017)
Fuertes G, Banterle N, Ruff KM, Chowdhury A, Mercadante D, Koehler C, Kachala M, Estrada Girona G, Milles S, Mishra A, Onck PR, Gräter F, Esteban-Martín S, Pappu RV, Svergun DI, Lemke EA
|
| RgGuinier |
2.4 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
16 |
nm3 |
|
|